Question
Organisms often release chemicals when attacked as part of their defence system. Scientists studied lima bean plants (Phaseolus lunatus) infested with either an armyworm, Spodoptera exigua, or a herbivorous mite, Tetranychus urticae. Both organisms feed on lima bean leaves, causing the leaves to release chemicals.
The study was conducted to see which defence chemicals were produced by lima bean leaves when infested by armyworms or herbivorous mites. The scientists identified a mixture
of compounds (C) released by the plant when attacked. Two of the chemicals in this mixture were identified (C1 and C2). The scientists hypothesized that the defence chemicals in C act as signals to produce other chemicals (X, Y and Z) that are also involved in the defence of the plant.
The graphs show the amounts of chemicals X, Y and Z produced when the plants were infested by either one of the two herbivores or treated with the different chemicals C1 or C2.
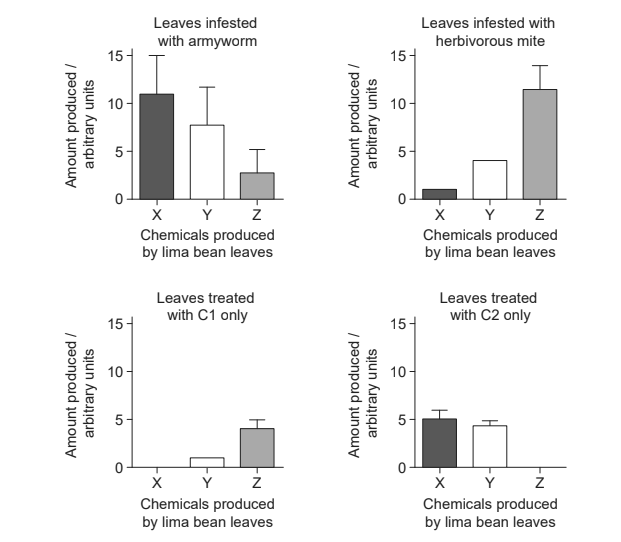
(a) Distinguish between the data for the leaves infested with the armyworm and the leaves infested with the herbivorous mite.
(b) Compare and contrast the effects of treatment of the leaves using C1 and C2 with the effects of infestation.
RNA was collected from leaves of the plants after each treatment (armyworm, herbivorous mite and the chemicals C1 and C2). DNA copies of the extracted RNA were made by a process called reverse transcription. Targeted genes in the DNA were then amplified.
(c) Identify the process that was used to amplify the targeted genes.
The scientists then used the transcribed DNA of each treatment to study the activation of three genes of the plants’ defence system. The DNA was separated by gel electrophoresis. The activation was tested one hour after treatment and again after 24 hours.
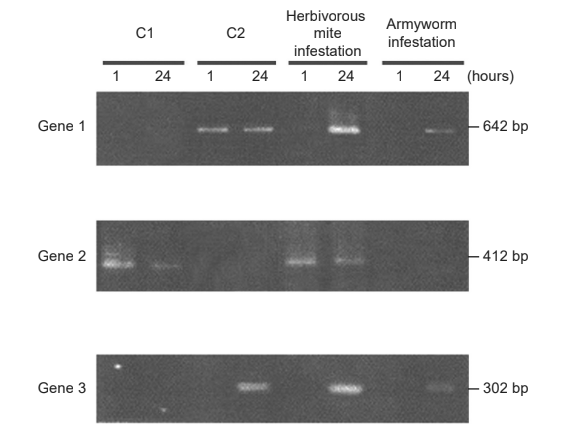
(d) Deduce, with a reason, which gene is first transcribed when exposed to C2.
(e) Each gene is activated by one or more of the treatments. From the gel electrophoresis data, discuss the impact of the herbivorous mite infestation on gene activation compared to treatment with C1 and C2.
(f) Using the gene activation data, deduce, giving two reasons, whether the armyworm or the herbivorous mite has infested lima bean plants over a longer period of time.
Answer/Explanation
Ans:
a)
a. armyworm «infestation» produced more X than Y than Z/decreasing amounts AND herbivorous mite showed the opposite pattern/more Z than Y than X
b. armyworm «infestation» produced more X than herbivorous mite
c. armyworm «infestation» produced more Y than herbivorous mite / Y is the middle value for both
d. armyworm «infestation» produced less Z than the herbivorous mite
e. other valid distinction
b)
a. C1 caused the leaf to produce two of the same chemicals/Y and Z as the attack of herbivorous mites in a similar pattern «but in lower quantities»
b. C1 produces the least «total» amount of chemicals of all the treatments
c. C2 has very similar pattern to those caused by the armyworms «but in lower quantities»
d. both herbivores caused a greater production of chemicals/all three chemicals compared to either C1 or C2
e. armyworms cause the greatest total amount of chemical production of any of the other treatments
f. other valid comparison of chemical effect versus herbivore effect
c) PCR Accept RT PCR.
d) gene 1 is first transcribed «after C2 treatment» as it shows activation after one hour
e )
a. herbivorous mites induce activation of gene 2 first «at 1 hour» and then also gene 1 and gene 3 «at 24 hours»
OR
herbivorous mite «infestation» is the only treatment to affect all three genes/leads to greater gene expression overall
b. gene 2 activation similar for mite and C1 «at both 1 and 24 hours»
c. gene 3 activation similar for mite and C2 «both at 24 hours»
d. gene 1 activation slower for mite compared to C2 but more intense (than C2 at 24 hours)
e. gene 1 and gene 3 expressed in higher amounts «after 24 hours» in mite infestation compared to C2 Both parts OWTTE required for mpd.
f)
a. the greater «gene expression» response of the lima bean plant to the mite infestation indicates longer evolutionary relationship
b. herbivorous mites cause more genes to be expressed/higher intensity of gene activation
Question
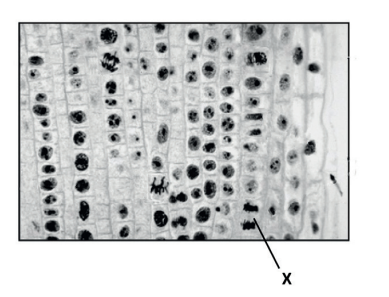
(a) (i) Identify the stage of mitosis labelled X in the image, giving a reason.
(ii) Outline what is indicated by the mitotic index of tissue taken from a tumour.
(iii) DNA has regions that do not code for proteins. State two functions of these regions.
1.
2.
(b) DNA methylation has a critical role in gene regulation by affecting transcription. Samples were taken from two colon cancer tumours (T1 and T2) and two normal colon samples (N1 and N2). A particular gene was implicated as a possible cause of cancer. The promoter of this gene was cloned (A–J). The data show the DNA methylation patterns from these samples. The numbers (32–269) represent different markers in the promoter.
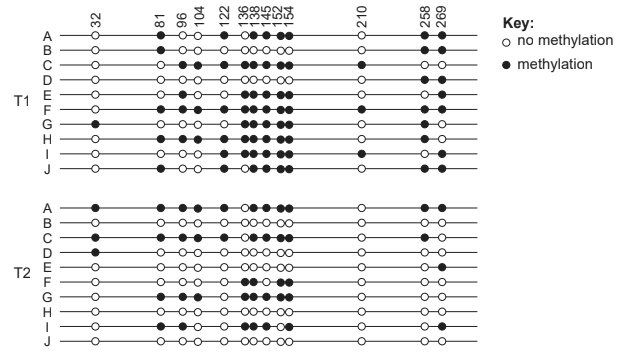
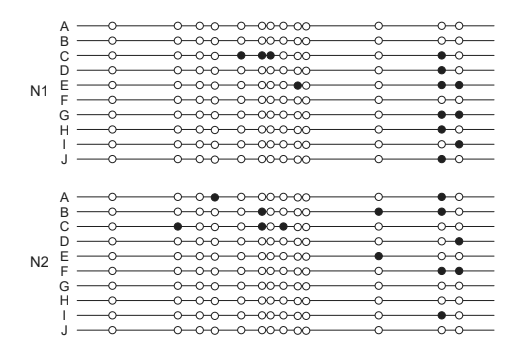
b(i) Outline the difference in methylation pattern between tumorous and normal tissue samples.
b(ii) Suggest a way methylation may affect tumour cell genes.
Answer/Explanation
Ans:
a (i) telophase because the chromosomes/chromatids have reached the poles
OR
«late» anaphase as some chromosomes/chromatids are still moving/tails visible OWTTE
a (ii)
a. mitotic index is an indication of the ratio/percentage of cells undergoing mitosis/cell division
b. cancer cells «generally» divide much more than normal «somatic» cells
c. a high/elevated mitotic index in tumours / possible diagnosis of cancer / measure of how aggressive/fast growing the tumour is
a (iii)
a. promoters / operators / regulation of gene expression/transcription
b. telomeres/give protection to the end of chromosomes «during cell division»
c. genes for tRNA/rRNA production
d. other valid function for non-coding sequence
b(i)
a. «overall» much more methylation in the colon tumour samples than normal
b. tumour and normal samples the markers 258 and 269 similar degree of methylation/fewer differences
c. degree of methylation on certain markers may correlate with the presence of cancer / correct example of a marker only methylated in tumour cells eg marker 32
b (ii)
a. «DNA» methylation may inhibit transcription of genes that would prevent cancer/tumor formation
