C. DNA Replication
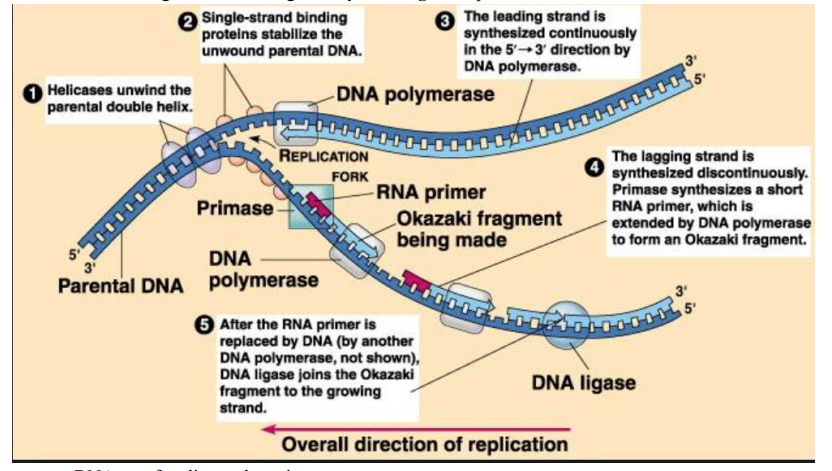
➢ First step is to unwind the double helix by breaking the hydrogen bonds
- Accomplished by an enzyme helicase
- Single stranded Binding Protein hold the strands apart
- Origin of replication=place where replication process begins; short stretch of DNA with specific nucleotide sequence
- Exposed DNA now forms a y-shaped replication fork
➢ DNA replication begins at specific sites called origins of replication
➢ Topoisomerase cuts and rejoins the helix to prevent tangling and relieve tension
➢ DNA polymerase III adds nucleotides to freshly built strand
- Can only add nucleotides to the 3’ end
➢ RNA primase adds a short strand of RNA nucleotides called an RNA primer
- Primase synthesizes RNA primer
- After replication, the DNA Polymerase I removes the RNA primer and replaces it with DNA
➢ Leading strand
- Synthesized continuously
- $5’ to 3’$
- Replicated towards fork
➢ Lagging strand
- Made in pieces called Okazaki fragments
- $3’ to 5’$
- Replicated Away from fork
- Must be made in pieces since nucleotides can only added to 3’ end
- Fragments linked together by DNA ligase to produce a continuous strand
➢ DNA proofreading and repair
- DNA polymerase in charge of repair synthesis
- Nuclease removes damage
- Ligase seals newly repaired strands
- Repair enzymes detect damage
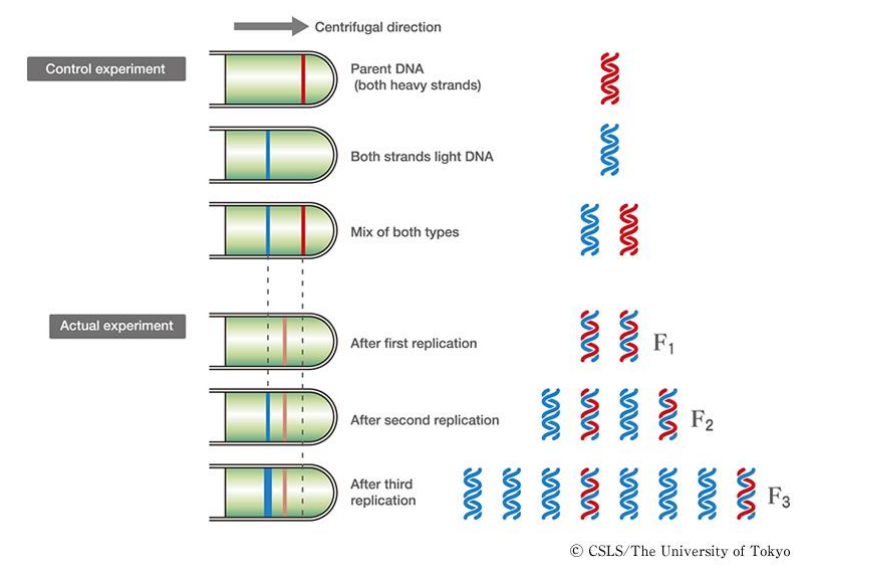
➢ Replicates semiconservatively because each new molecule is comprised of ½ of the original strand
- Semiconservative model proved by Meselson and Stahl’s experiments
■ Created “heavy” template DNA using N15, measured weights of replicated DNA by looking at the layers that formed, semiconservative model was the only one that fit
➢ A few bases at very end cannot be replicated because the DNA polymerase needs to bind
- Every time replication occurs the chromosome loses a few base pairs
- Genome has compensated for this over time by putting bits of unimportant/less important DNA at the ends of a molecules called telomere
➢ Key history
- Protein originally thought to be the carrier of genetic material due to its higher variety and specific functions
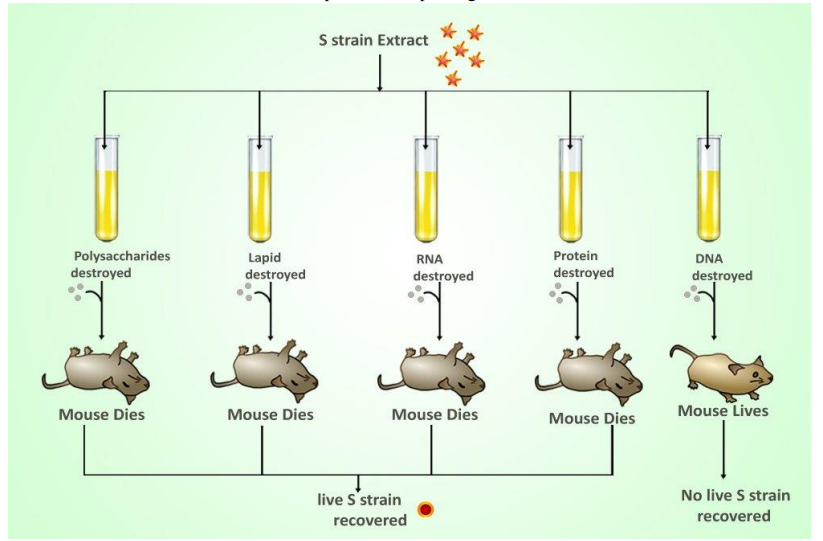
- Griffiths want to find out what substance causes transformation
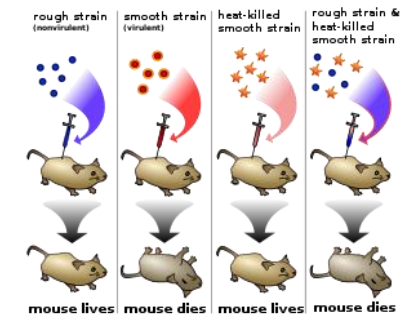
R bacteria had been transformed into pathogenic S bacteria by unknown substance
- A very, MacLeod, and McCarty isolated various cellular components from a dead virulent strain of bacteria
■ Followed up on a previous experiment by Griffiths and added each of these cellular components to a strain of living nonvirulent bacteria
■ Only the component of the deadly bacteria was able to change the second bacteria into a deadly strain capable of reproducing
■ DNA must be responsible for passing traits and it is inheritable
- Hershey-CHase
■ Used bacteriophages and labelled protein parts of some with radiolabeled sulfur and labelled the DNA parts of other viruses with radiolabeled phosphorus
■ Bacteriophages inject genetic material into cell so more genetic material will be created
■ When viruses infected bacteria, only the labelled DNA was inside, but they were still able to replicate and make progeny viruses
■ E. coli were infected by the phage, and there was more and more P that entered. They concluded that DNA carried the genetic information to produce DNA and proteins
➢ Central Dogma
- DNA’s main role is directing the manufacture of molecules that actually do the work in the body
- DNA expression:
- 1. Turn into RNA
- 2. Send RNA out into the cell and often gets turned into a protein
➢ Transcription turns RNA into DNA
- Takes place in nucleus (except in prokaryotes)
➢ Translation turns RNA into a protein
- Takes place in cytoplasm
➢ RNA
- Single stranded
- 5-carbon sugar is ribose instead of deoxyribose
- Uses uracil instead of thymine
- major types of RNA
■ Messenger RNA (mRNA)
● Temporary version of DNA that gets sent to ribosome
■ Ribosomal RNA (rRNA)
● Produced in nucleolus
● Makes up part of ribosomes
■ Transfer RNA (tRNA)
● Shuttles Amino acids to the ribosomes
● Responsible for bringing the appropriate amino acids into place at the appropriate time
● Done by reading message carried by mRNA
■ Interfering RNA (RNAi)
● Small snippets of RNA that are naturally made in the body or intentionally created by humans
● siRNA and miRNA can bind to specific sequences of RNA and mark them for destruction
➢ Transcription
- RNA copy of DNA code
■ pre-mRNA synthesis - Only a specific section is copied into mRNA
- Occurs as-needed on a gene-by-gene basis
- Exception: prokaryotes will transcribe a recipe that can be used to make several proteins
■ Called polycistronic transcript
■ Eukaryotes tend to have one gene that gets transcribed to one mRNA and translated into one protein
● Monocistronic transcript
- 3 steps: initiation, elongation, termination
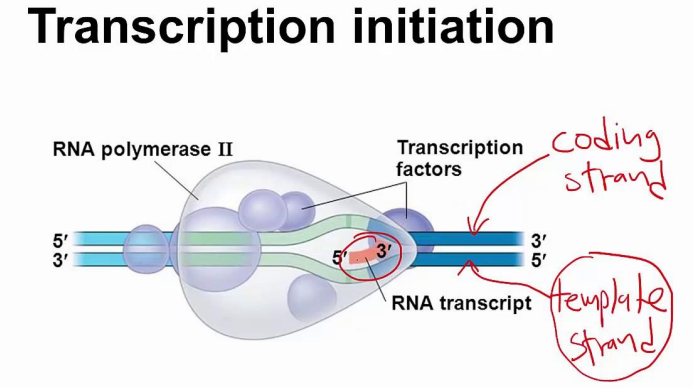
■ initiation
● Unwind and unzip DNA strands using helicase
● Transcription initiation complex - Transcription factor proteins+RNA polymerase
- Forms at promoter
● Transcription only occurs as-needed to conserve resources
■ ELongation
● Begins at special sequences of the DNA strand called promoters
● Free RNA nucleotides inside the nucleus used to create mRNA - RNA polymerase used to construct mRNA
● Strand that serves as the template is called antisense strand, the noncoding strand, or the template strand
● Strand that lies dormant is the sense strand, or the coding strand
● Rna polymerase build RNA only to 3’ side - Doesn’t need primer
● Promoter region is “upstream” of the actual coding part of the gene
● Official starting point if start site
● RNA strand is complementary to template DNA strand
■ Termination
● Once termination sequence is reached, it separates from the DNA template, completing the process of transcription
➢ RNA processing
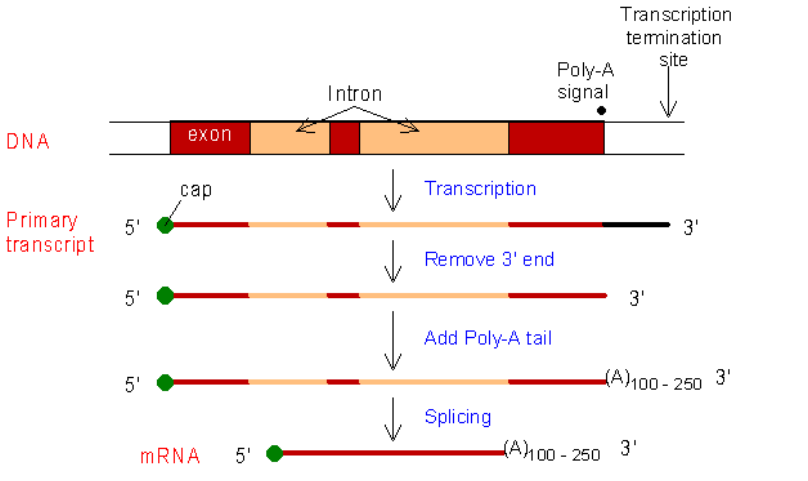
- In eukaryotes the RNA must be processed before it can leave the nucleus
- Freshly transcribed RNA is called hnRNA (heterogenous nuclear RNA) and it contains both coding regions and noncoding regions
- Regions that express the code will be turned into protein are exons
- Non-coding regions in the mRNA are introns
■ Introns removed by spliceosome
● Spliceosome made up of many snRNPs - snRNPs made up of ribozyme+small nuclear RNA
■ ribozyme=RNA catalyst that can copy RNA strands
● Spliceosome identifies ends of an intron
● Folds chromosome
● Spliceosome cuts out the intron and binds the two exons together
● Non i prokaryotic cells
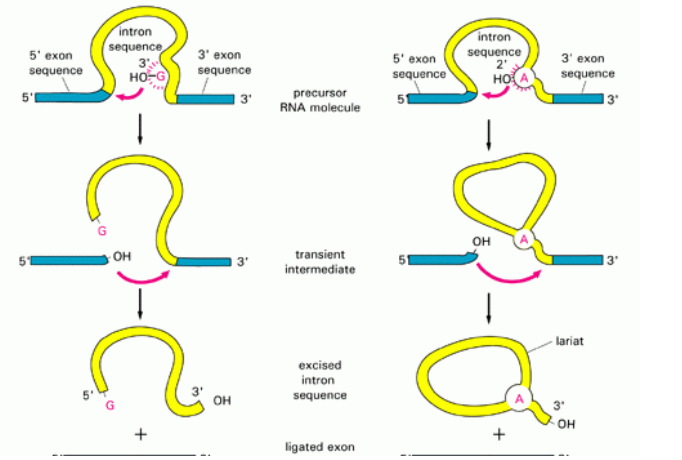
● 3 properties of RNA that allow it to function as an enzyme
- Single stranded
- Functional groups that act as catalysts
- Hydrogen bonds with other nucleic acids
■ Introns allow more genetic diversity
● More possibilities for crossover
● Alternate splicing can yield new protein varieties - Methyl cap added to 5’ end
■ Helps mRNA leave nucleus
■ Allows attachment to ribosome
■ Modified guanine
○ poly-A tail added to 3’ end
■ Protects mRNA from endonucleases in cytoplasm, which can only attach to 3’ end
■ 50-250 adenine
○ No cap or tail in prokaryotic cells since they have no nucleus
○ mRNA leaves nucleus through nuclear pore
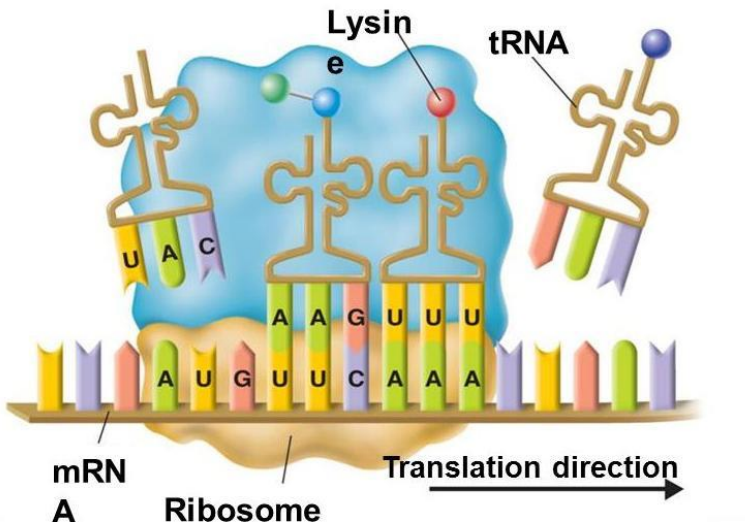
➢ TRanslation
- Process of turning mRNA into a protein
- mRNA nucleotides will be read in the ribosome in groups of three
■ Group of three nucleotides=codon
● Each codon corresponds to a particular amino acid - mRNA attaches to the ribosomes to initiate translation and “waits” for the amino acids to come to the ribosome
- 3 sites on ribosome
■ E=exit site
■ P=polypeptide storage/exit
■ A= place where tRNA brings in amino acid - 1.mRNA attaches to mRNA binding site on small subunit. tRNA attaches onto A site
- 2. Large subunit attaches via GTP
■ 1st tRNA is in P site
■ 2nd tRNA comes in A site - 3. rRNA in large subunit catalyzes a peptide bond between amino acids
○ 4. 1st rRNA moves to exit site
■ 2nd tRNA moves to P site
■ New tRNA comes in through A site
■ Steps 1-4 repeats until stop codon is reached
■ As mRNA moves through ribosome, other ribosomes can attach to it at the same time (as long as mRNA has not degraded, especially on 5’cap
- 5. Release factor adds water to end of polypeptide; polypeptide detaches and exits
through P-site tunnel - 6. small/large subunit/mRNA disassemble and disassociate; process of translation can start over again
- tRNA carries amino acid
■ Attaches to RNA via anticodon (complementary base pair to codon)
■ Wobble pairing on third nucleotide (flexible bonds)
■ Each tRNA becomes charged and enzymatically attaches to an amino acid in the cell’s cytoplasm and “shuttle” it to the ribosome
■ Charging enzymes require ATP - 3 phases:
■ Initiation
● 3 binding sites: - A site
- P site
- E site
● Start codon is AUG (methionine)
● TATA box=specific promoter for initiation
● RNA polymerase binds to a specific location on promoter
● Transcription factors attach to promoter to help guide RNA polymerase
■ Elongation
● As each amino acid is brought to the mRNA, it is linked to its neighboring amino acid by a peptide bond and eventually forms a full protein
■ Termination
● Synthesis of a polypeptide ended by stop codons
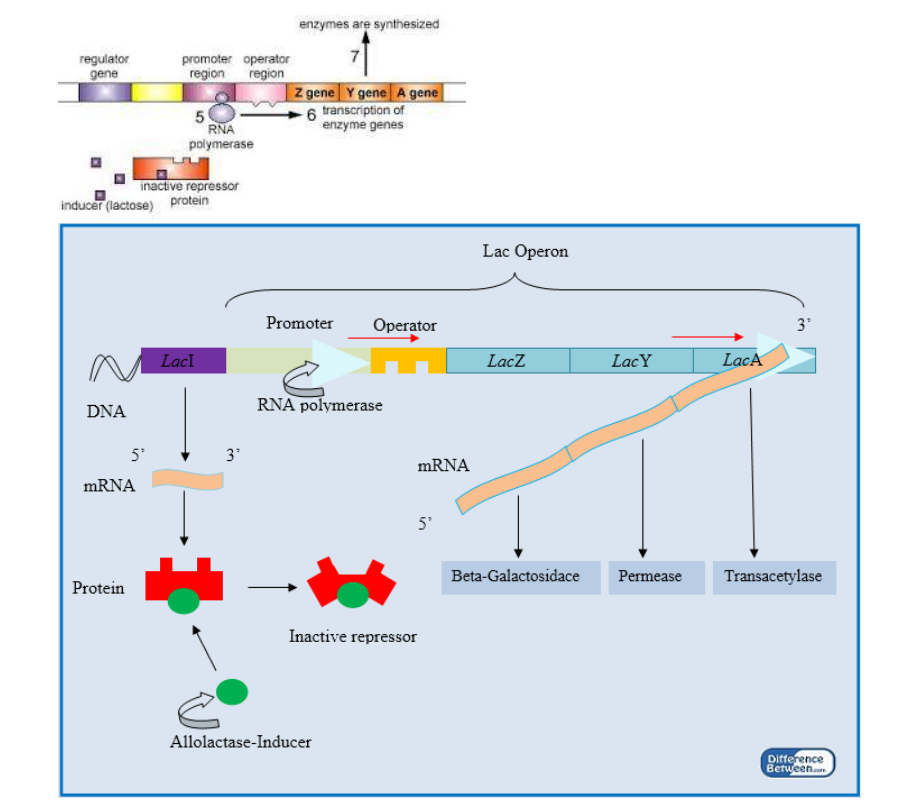
➢ Gene Regulation
- Pre-transcriptional regulation
■ Transcription factors can either encourage or inhibit the unwinding of DNA and the binding of RNA polymerase - Operons
■ Bacteria only
■ Structural genes
● Code for enzymes in a chemical reaction
● Genes will be transcribed at the same time to produce particular enzymes
■ Promoter gene
● Region where RNA polymerase binds to begin transcription
■ Operator
● Controls whether transcription will occur
● Where repressor/inducer binds
■ Regulatory gene
● Codes for a specific regulatory protein called to repressor
● Repressor capable of attaching to the operator and blocking transcription
● If repressor binds to the operator, transcription will not occur
● If repressor does not bind to the operator, RNA moves along operator and transcription occurs
■ inducible=presence of molecule turns gene on
■ repressible=presence of molecule turns gene off
- Chromatin Modification
■ Histone acetylation
● Acetyl groups added to histones
● Looser Chromatin
● Access for transcription increased
■ DNA methylation
● Methyl groups added to bases
● Silences gene
● Tightens chromatin - Enhancers
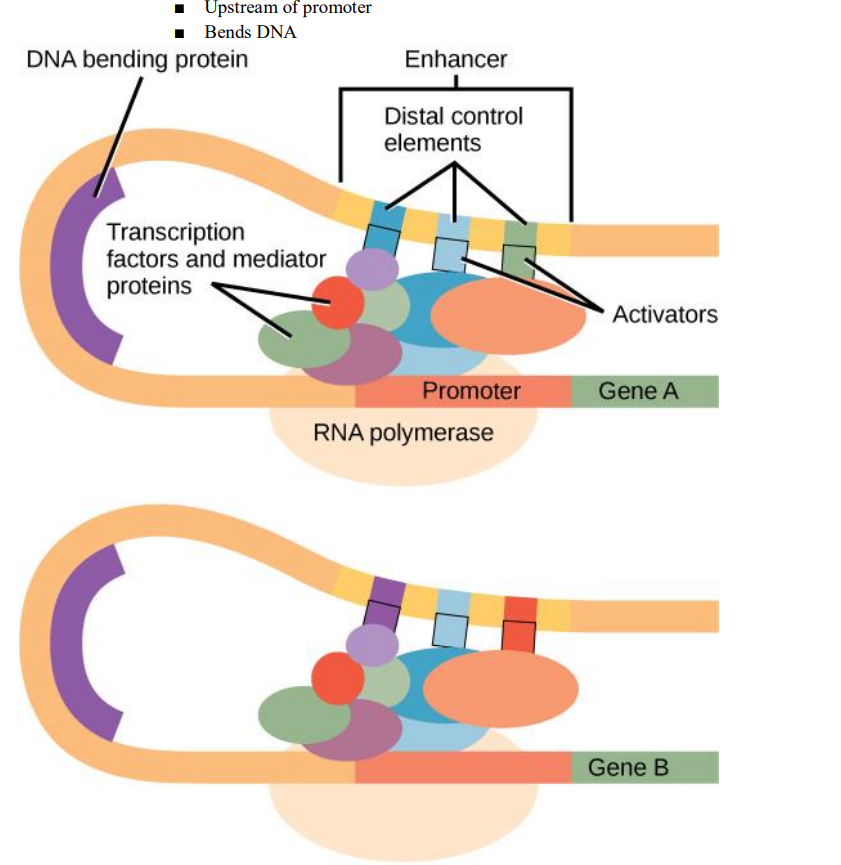
- Post-transcriptional regulation
■ Occurs when the cell creates an RNA but then decides that it should not be translated into a protein
■ RNAi molecules bind to an RNA via complementary base pairing
■ Creates double-stranded RNA, signalling that RNA should be destroyed, preventing it from being translated - Post-translational regulation
■ Protein has already been made, but doesn’t need it yet, so it is deactivated
➢ Mutations
- Mutation is an error in the genetic code
- Occur because DNA is damaged in cannot be repaired or because DNA is repaired incorrectly
■ Caused by chemicals or radiation
■ Can also occur when a DNA polymerase or an RNA polymerase makes a mistake
■ RNA polymerases have proofreading abilities, but RNA polymerases do not - Error in DNA not a problem unless that gene is expressed AND the error causes a change in the gene product
➢ Base Substitution
- Point mutations result when a single nucleotide is replaced for another
- Nonsense mutation
■ Cause original codon to become a stop codon, which results in early termination of protein synthesis - Missense mutation
- Cause original codon to be altered and produce a different amino acid
- Silent mutations
■ codon that codes for the same amino acid is created and therefore does not
change the corresponding protein sequence - Frameshift mutations
■ insertions/deletions result in the gain or loss of DNA or a gene
● Can have devastating consequences during translation - results in a change in the sequence of codons used by the ribosome
- Duplications
■ Extra copy of genes
■ Caused by unequal crossing-over during by meiosis or chromosome
rearrangements
■ ,ay result in new traits as one copy may evolve a new function
○ Inversions
■ Changes occur in the orientation of chromosomal regions
■ May cause harmful effects if the inversion involves a gene or an important regulatory sequence - Translocation
■ 2 different chromosomes break and rejoin in a way that causes the DNA sequence to be lost, repeated, or interrupted