Topic 7: Nucleic acids
7.1 DNA structure and replication
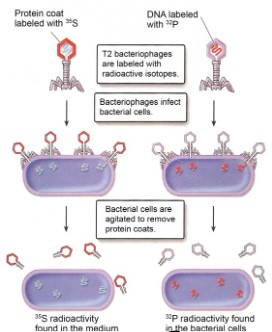
Hershey & Chase experiment
Experiment target: T2 Bacteriophage (a kind of virus, containing DNA and a protein coat) & E.Coli bacteria
How does virus work?
- Virus injects their genetic material into cell
- Non-genetic part of the virus (protein coat) remains outside the cell
- Infected cell produces large amount of viruses based on the genetic material in the cell
- The cell bursts releasing copied viruses
Experimental methodology:
- Amino acid containing radioactive isotopes were used to label the virus: \(^{35}S\) is used for labelling the protein coat, and \(^{32}P\) is used for labelling DNA
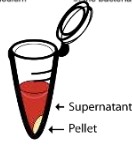
- Centrifuge is used for vigorous shaking in order to separate virus and infected bacteria:
Pellet (solid) contains bacteria and supernatant (liquid) contains protein coat.
Findings:
- When protein coat is only radioactively labelled, new viruses made shown no radioactive sign
- When DNA is only radioactively labelled, new viruses made shown radioactive sign
Hypothesis: DNA controls genetic information
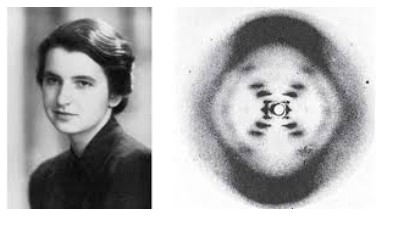
Rosalind Franklin’s and Maurice Wilkins’ investigation of DNAs tructure by X-ray diffraction
Rosalind Franklin is the first women to take a photo of DNA using x-ray diffraction. Her
investigation leads to the findings that DNA is a double helix.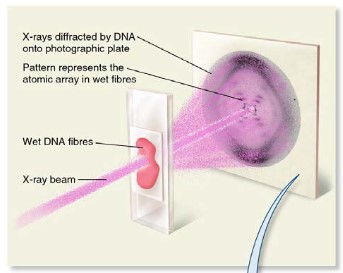
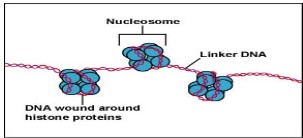
Nucleosome
- Eukaryotic DNA supercoiling is organized by nucleosome
- Nucleosomes are formed by wrapping DNA around histone protein
- Supercoiling in general helps regulate transcription because only certain areas of the DNA are accessible for the production of mRNA by transcription. This regulates the production of a polypeptide.
- Nucleosomes will be further supercoiled to form chromosome, which is only formed in cell division.
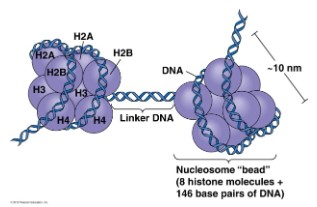
DNA will wrap around the histone protein (as shown as the blue ball in the diagram)
first, histones will then stick together to form nucleosome (8 histone protein). Only the
DNA connecting two nucleosome (Linker DNA in the diagram) is accessible and can
be transcribed into mRNA to produce polypeptides
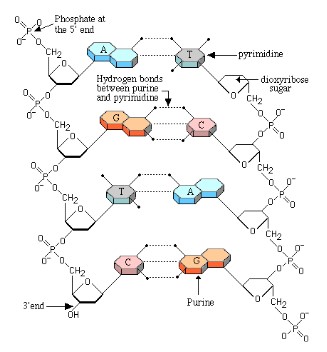
DNA Structure
- DNA is double stranded and shaped like a ladder, with the sides of the ladder made out of repeating phosphate and deoxyribose sugar molecules covalently bonded together.
- Each deoxyribose molecule has a phosphate covalently attached to a 3’ carbon and a 5’ carbon.
- The phosphate attached to the 5’ of one deoxyribose molecule is covalently attached to the 3’ of the next deoxyribose molecule forming a long single strand of DNA known as the DNA backbone.
- DNA strands run anti-parallel to each other with one strand running in a 5’ to 3’ direction and the other strand running 3’ to 5’ when looking at the strands in the same direction
- The rungs of the ladder contain two nitrogenous bases (one from each strand) that are bonded together by hydrogen bonds.
- Since these two strands are anti-parallel replication occurs in different directions on the DNA strand
- Purines are two ring nitrogenous bases and pyrimidines are single ring nitrogenous bases.
- Adenine and thymine has two hydrogen bonds; Guanine and Cytosine has three hydrogen bonds
DNA replication
- DNA replication creates two identical strands with each strand consisting of one new and one old strand (semi-conservative).
- DNA replication occurs at many different places on the DNA strand called the origins of replication (represented by bubbles along the strand).
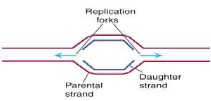
Several site “replication bubbles” starts together, so replication process is much faster. Replication starts
at the replication forks
- The DNA strand is unwound and separated (hydrogen bonds broken) by an enzyme called helicase

- DNA gyrase is an enzyme that relieves strain on the strand as it is being unwound by the helicase
- Single-stranded binding proteins bind to the open strands and keep the strands apart long enough to prevent the strands from re-annealing before the strands are copied
- RNA primase attaches a RNA primer to the DNA as starting point and attachment point since DNA polymerases can only add nucleotides to the 3’ end of a primer
- DNA polymerase III adds free nucleotides found in the nucleus in the 5’ to 3’ direction in the direction of the replication fork.
- This strand is called the “leading strand” because replication is continuous (keeps replicating until it meets another origin of replication).
- Because DNA polymerase III can only add nucleotides to a free 3’ end (5’to 3’ direction) the other strand is replicated in the opposite direction.
- This strand is called the “lagging strand” because replication is delayed until the helicase opens up available nucleotides, to allow replication “back” in the opposite direction to the leading strand. Replication in this direction is discontinuous.
- The lagging strand is therefore made as a series of fragments called “Okazaki fragments”
- DNA polymerase I replaces RNA primer with DNA
- DNA ligase glues the Okazaki fragments together to form a continuous strand
Non-coding genes
- Genes contained within DNA called coding sequences, code for polypeptides created during transcription and translation
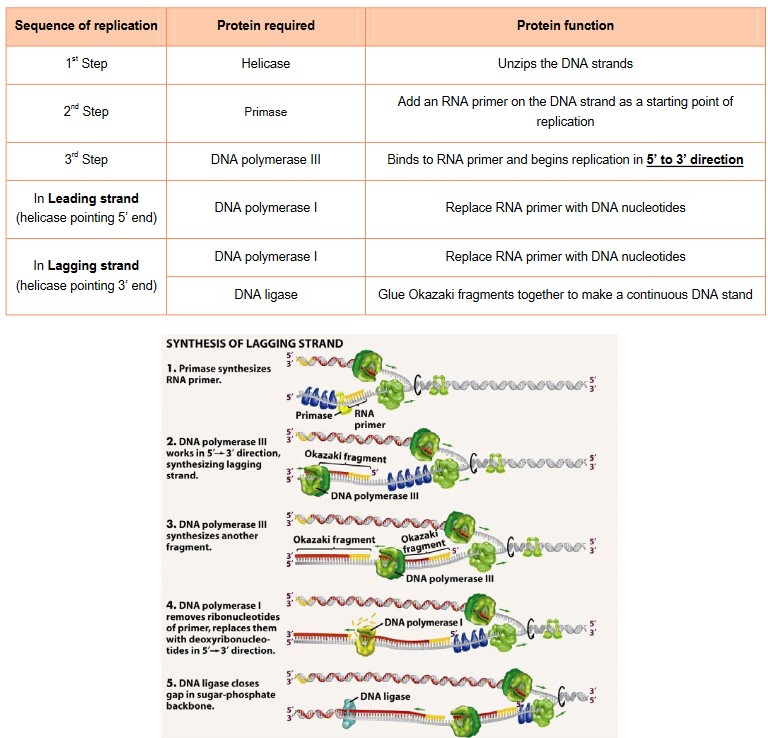
- The majority of DNA are non-coding sequences that perform other functions such as regulators of gene expression, introns, telomeres.
- Silencers are DNA sequences that bind regulatory proteins called repressors that inhibit transcription
- Promoters are the attachment points for RNA polymerase to transcribe mRNA
- Enhancers are DNA sequences that increase the rate of transcription
- Introns are the area in DNA for non-coding regions
- Extrons are the area in DNA for coding regions
- mRNAonly copies the information from the extrons and ignore what is in the introns. So only extrons’ information can leave the nucleus
- In DNA there are also many repetitive sequences, especially in eukaryotic DNA, that can make up 5-60% of the genome; specifically, an area of repetitive sequences that occurs on the ends of eukaryotic chromosomes.
- These repetitive sequences called telomeres, protect the DNA during replication. Since enzymes can’t replicate all the way to the end of the chromosome, the parts that aren’t copied are part of the telomeres. This prevents the DNA molecule from degradation during replication.
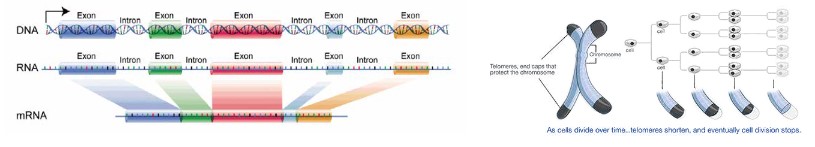
Artificial DNA sequencing
Dideoxyribonucleotides (DDNA) inhibit DNA polymerase during replication, thereby stopping replication from continuing. Dideoxyribonucleotides with fluorescent markers, are used and incorporated into sequences of DNA, to stop replication at the point at which they are added. This creates different sized fragments with fluorescent markers that can be separated and analyzed by comparing the colour of the fluorescence with the fragment length
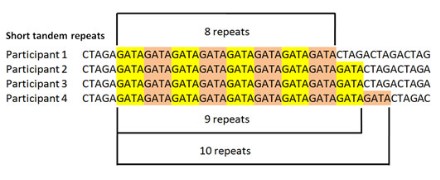
Tandem repeat
Short tandem repeats (STRs), also known as variable tandem repeats (VNTRs) are regions of non-coding DNA that contain repeats of the
same nucleotide sequence. They are repeated numerous times in a head-tail matter. These short repeats show variations between individuals in terms of the number of times the sequences is repeated.
For example, CATACATACATACATACATACATA is a STR where the nucleotide sequence CATA is repeated six times for one individual. However, in another individual, this tandem repeat could occur only 4 times CATACATACATACATA. These variable tandem repeats are the basis for DNA profiling used in crime scene investigations and genealogical tests (paternity tests).
Paternity test: Mother’s DNA – mitochondrion
Father’s DNA – Y chromosome