Topic 7: Nucleic acids
7.2 Transcription and gene expression
Transcription of RNA
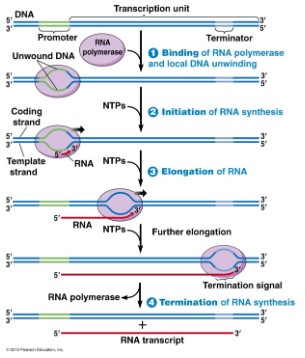
- Transcription occurs in a 5’ to 3’ direction where the 5’ end of the free RNA nucleotide is added to the 3’ end of the RNA molecule that is being synthesized.
- Transcription consists of 3 stages called initiation, elongation and termination
- Transcription begins when the RNA polymerase binds to the promoter with the help of specific binding proteins
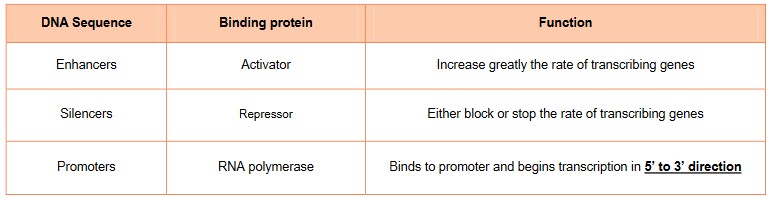
The promoter region is a DNA sequence that initiates transcription and is an example of
non-coding DNA that plays a role in gene expression. Promoter region is also the binding site for
RNA polymerase, is the starting point of transcription
Operator allows inhibitor protein to bind to stop the transcription, Repressor protein is bind to the
operator to prevent RNA polymerase from transcribing genes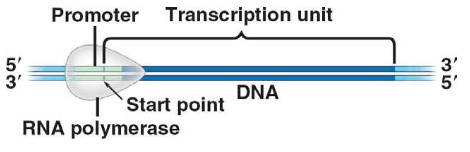
Nucleosome regulating gene transcription
- As explained previously in 7.1 eukaryotic DNA wraps around histone proteins and supercoils
- This supercoiling helps regulate transcription because only certain areas of the DNA are accessible for the production of mRNA by transcription.This regulates the production of a polypeptide.
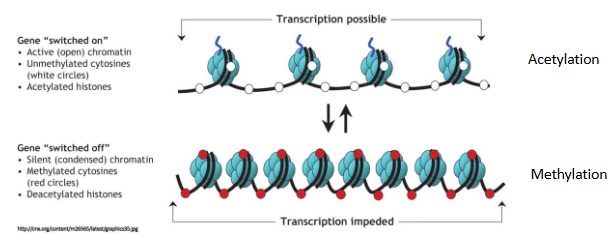
- One of the main ways this occurs is through the modification of the histone tails
- Acetylation: When acetyl groups are added to the positively charged histone tails, they become negative and the DNA repels against them. This opens up the nucleosome so the DNA is not as close to the histone anymore and chromatin remodeling can occur. Acetylation
switches on genes. - Methylation: decreases transcription of the gene. It makes histone protein closer and tighter so less gene will be expressed. Methylation switches off genes.
- The amount of methylation can vary over an organisms lifetime and can be affected by environmental factors
Protein regulation of gene expression
- Gene expression can also be regulated by the environment surrounding the gene that is expressed or repressed
- Specific proteins can regulate how much transcription of a particular gene will occur
- These regulatory proteins are unique to a particular gene
- Regulatory sequences on the DNA that increase the rate of transcription when proteins bind to them are called enhancers
- Regulatory sequences on the DNA that decrease the rate of transcription when proteins bind to them are called silencers
- Promoter-proximal elements have binding sites closer to the promoter and their binding is necessary to initiate transcrip
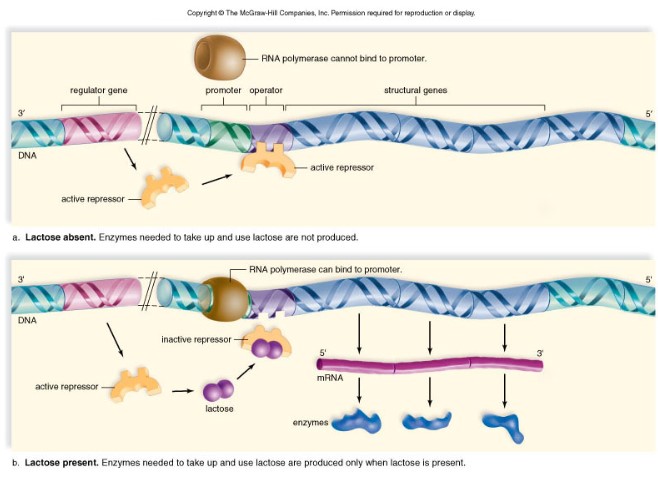
Example: Lactose production
- In prokaryotic cells such as E.coli repressor proteins block the production the enzymes needed to break down lactose in the cell.
- However, when Lactose is present, it will bind to the repressor protein, causing it to fall off, and allowing transcription to occur.
- As transcription occurs, these enzymes are made and lactose is broken down into glucose and galactose. Since there is small amounts of lactose now in the cell, the repressor binds again to the operator, blocking transcription from taking place.
- This is an example of negative feedback
Environmental impact on gene expression
- The external environment in which the organism is located or develops, as well as the organism’s internal world, which includes such factors as its hormones and metabolism can have an impact on gene expression
- Temperature and light are external conditions which can affect gene expression in certain organisms.
- For example, Himalayan rabbits carry the C gene, which is required for the development of pigments in the fur, skin, and eyes, and whose expression is regulated by temperature (Sturtevant, 1913).
- Specifically, the C gene is inactive above 35°C, and it is maximally active from 15°C to 25°C. This temperature regulation of gene expression produces rabbits with a distinctive coat coloring. In the warm, central part of the rabbit’s body, the gene is inactive, and no pigments are produced therefore the fur color is white (picture below). In the rabbit’s extremities (i.e., the ears, tip of the nose, and feet), where the temperature is much lower than 35°C, the C gene actively produces pigment, making these parts of the animal black.

- During embryonic development embryos contain chemicals called morphogens, which can affect gene expression and thereby affecting the fate of embryonic cells depending on their position within the embryo. Morphogens regulate the production of transcription factors in a cell
- An obvious example is how sunlight affects the production of skin pigmentation in humans
- A chemical example, was the use of Thalidomide by pregnant woman for morning sickness. It was thought it was harmless for humans but was not thoroughly tested. The drug was withdrawn too late to prevent severe developmental deformities in approximately 8,000 to 12,000 infants, many of whom were born with stunted limb development. Interestingly, despite the fact that thalidomide is dangerous during embryonic development, the drug continues to be used in certain instances yet today.
Modification of mRNA after transcription
- In eukaryotes, the locations for transcription and translation are separated by the nuclear membrane. This allows for post-transcriptional modification of the mRNA.
- The first product of transcription is pre-mRNA
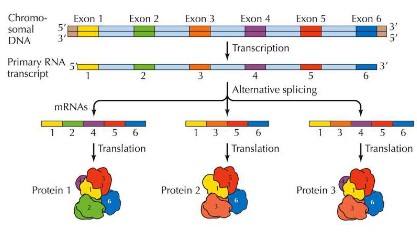
- As eukaryotic mRNA travels from the nucleus to the ribosomes, non-coding strands of the mRNA called introns are removed to form functional mature mRNA.
- They are removed through RNA splicing
- The exons are spliced together to form mature mRNA
- Also a poly A tail consisting of approximately 100-200 adenine nucleotides is added to one end of the mRNA and a 5’ cap is added to the other end (these help protect the mature mRNA transcript)
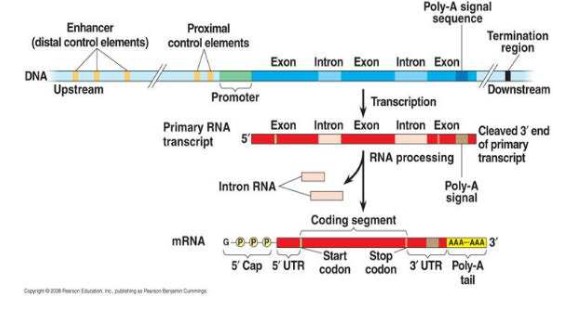
- Alternative splicing can also occur with genes that produce multiple proteins, which means that some exons may also be removed during splicing, thus producing different polypeptides
- For example, in mammals tropomyosin which is a protein involved in muscle contractions; however, the pre-mRNA is spliced to form
5 different forms of the protein. The mature mRNA that codes for tropomyosin in the smooth muscle of the intestines is missing
exon 3 and 10,while the mRNA that codes for tropomyosin in skeletal muscle is missing exon 2.