Topic 7: Nucleic acids
7.3 Translation
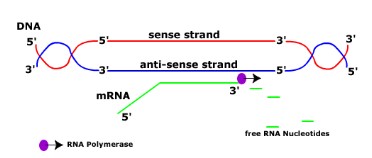
Transcription(revised)
- RNA polymerase binds to a site on DNA (promoter region) at the start of a gene.
- RNA polymerase separates the DNA strands and synthesises a complementary RNA strand.
- RNA copies from the anti-sense strand which contains the information on the sense strand.
- Once RNA is made, RNA polymerase detaches and DNA double helix reform
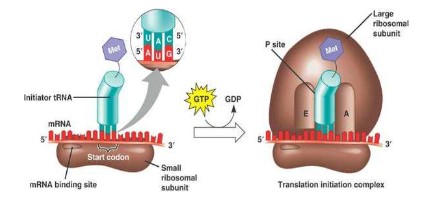
Initiation of translation:
- mRNA binds to the small (30 s) ribosomal sub-unit.
- tRNA carrying Methionine with the anticodon UAC binds to the codon AUG (start codon).
- This is called the initiation complex.
- The large ribosomal subunit binds to the small ribosome, with the tRNA containing methionine binding at the p-site of large subunit.
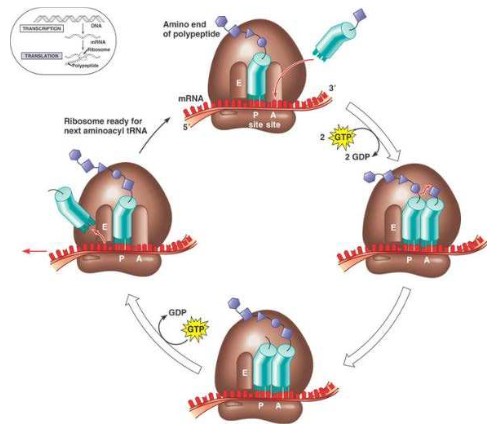
Elongation of translation
- While the first tRNA is still attached, a second tRNA attaches to the mRNA at the A site on the ribosome, carrying the amino acid that corresponds to the mRNA codon.
- The methionine amino acid at the P site binds to the amino acid carried by the second tRNA located at the A site.
- The two amino acids are joined together through a condensation reaction that creates a peptide bond between the two amino acids.
- The ribosome moves along the mRNA one codon shifting the tRNA that was attached to methionine to the E site.
- The tRNA is released back into the cytoplasm from the E site, allowing it to pick up another amino acid (methionine) to build another polypeptide.
- Another tRNA moves into the empty A site bringing the next amino acid corresponding to their RNA codon.
- Again, the amino acid is attached to the polypeptide forming a peptide bond, the ribosome slides across one codon and tRNA at the P site moves into the E site releasing it back into the cytoplasm.
- The ribosome continues to move along the mRNA adding amino acids to the polypeptide chain.
- This process continues until a stop codon is reached.
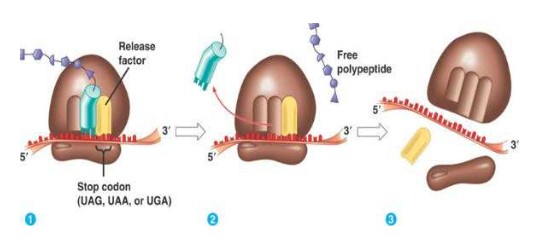
Termination of translation:
- Termination begins when 1 of the 3 stop codons (UAA, UGA, UAG) moves into the A site.
- These tRNA have no attached amino acids.
- When the stop codon is reached the ribosome dissociates and the polypeptide is released
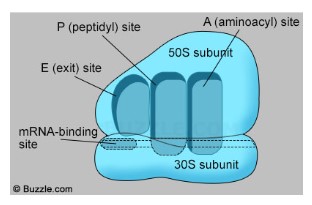
Ribosome
- Ribosomes are composed with two subunits – large and small subunit
- Large subunit sit on the top and small subunit holds the RNA
- Ribosomes have three sites:
- A site: check for the right tRNA matching mRNA, joining to the mRNA (Activation)
- P Site: form peptide bonds with growing amino acid chain (Polypeptide)
- E site: empty tRNA exit back to cytoplasm to pick up new amino acid (Exit)
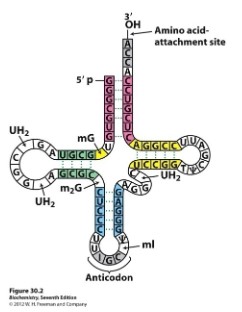
tRNA Structure
- tRNA is a type of RNA molecule that transfers a specific amino acid to a growing polypeptide chain during translation (protein synthesis) at the ribosomes.
- Sections of the tRNA become double stranded through hydrogen bonds formed between base pairs creating loops
- A triplet of bases form the anticodon which will bind to the corresponding triplet codon on the mRNA strand
- The base sequence of CCA at the 3’ end forms the amino acid binding site
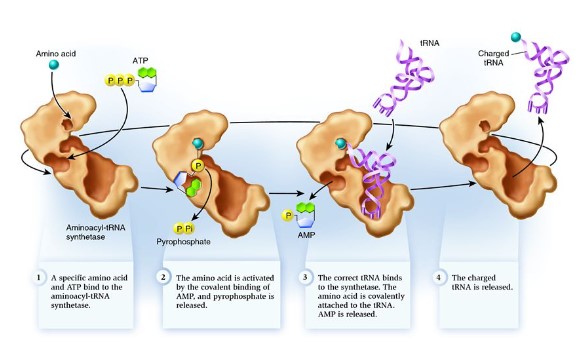
tRNA activating enzyme
- Each tRNA binds with a specific amino acid in the cytoplasm in a reaction catalyzed by a specific tRNA-activating enzyme.
- Each specific amino acid binds covalently to the 3′- terminal nucleotide (CCA) at the end of the tRNA molecule.
- The binding of the specific amino acid to the tRNA requires energy from ATP.
- Energy between the tRNA and amino acid (the bond) will be used in translation to form a peptide bond between amino acids
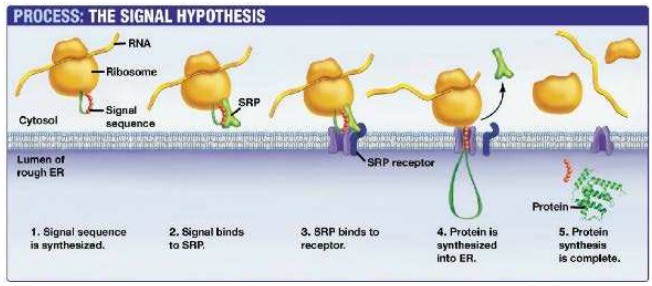
Ribosomes on rough endoplasmic reticulum synthesise protein for secretion
- Ribosomes attached to ER create proteins that are secreted from the cell by exocytosis or are used in lysosomes.
- Proteins perform many functions within specific compartments of the cell or in other parts of the body after they are secreted out of the cell
- Proteins that are destined to be used in lysosomes, ER, Golgi Apparatus, the plasma membrane or secreted by the cell are made by ribosomes bound by the endoplasmic reticulum
- Ribosomes that become bound to the ER are directed here by a signal sequence that is part of that specific polypeptide
- This signal sequence on the polypeptide binds to a signal recognition protein (SRP)
- The SRP guides the polypeptide and ribosome to the ER where it binds to an SRP receptor
- Translation can now continue and the polypeptide is deposited into the lumen of the ER as its created for transportation to the correct
location
Four levels of protein structure
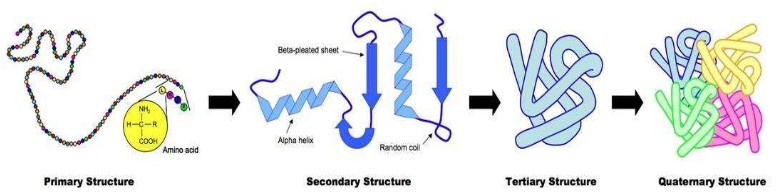
- Primary structure: basic amino acid chain
- Secondary structure: Held together by hydrogen bonds between (non-adjacent) amine (N-H) and carboxylic (C-O) groups, H-bonds provide a level of structural stability. (alpha helix shape & beta pleated sheet) (e.g. silk)
- Tertiary structure: The polypeptide folds and coils to form a complex 3D shape. Caused by interactions between R groups including ionic bonds, sulfur bridge, hydrophobic and hydrophilic interaction. Protein is globular in nature.
- Quaternary structure: 2 or more polypeptide chains and/or an inorganic compound (prosthetic group) (e.g. hemoglobin)