Principles and process of Biotechnology : Notes and Study Materials -pdf
Notes and Study Materials
- Concepts of Principles and process of Biotechnology
- Master File Principles and process of Biotechnology
- NCERT Book chapter Principles and process of Biotechnology
- NCERT Solutions for – Principles and process of Biotechnology
- NCERT Exemplar Solutions for – Principles and process of Biotechnology
- Revision Note of Principles and process of Biotechnology
- Past Many Years Question papers and Answer of Principles and process of Biotechnology
- Mind Map of Principles and process of Biotechnology
Examples and Exercise
11. BIOTECHNOLOGY: PRINCIPLES & PROCESSES
|
– The product is formulated with suitable preservatives. Such formulation undergoes thorough clinical trials and strict quality control testing
CBSE Class 12 Biology Important Questions Chapter 11 – Biotechnology: Principles and Processes
1 Mark Questions
Chapter 11
Biotechnology Principles and Processes
1 Marks Questions
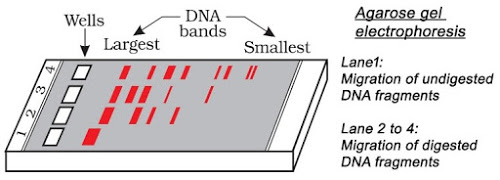
1. A restriction enzyme digests DNA into fragments. Name the technique used to check the progression of this enzyme and separate DNA fragments.
Ans. Gel electrophoresis
2. Name two commonly used vectors in genetic engineering.
Ans. Plasmid and Bacteriophage.
3. Some enzymes are considered as molecular scissors. in genetic engenrring. What is the name assigned to such enzymes?
Ans. Restriction Enzymes.
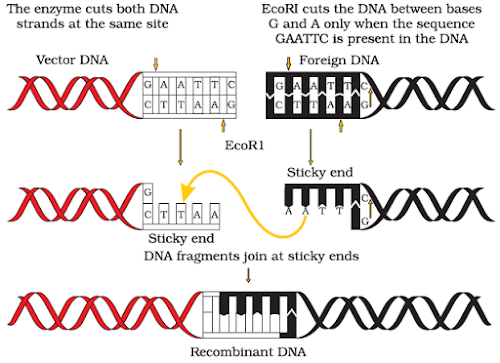
4. Write conventional nomenclature of EcoRI.
Ans.E. – Escherichia; co – coli; R – Name of Strain; I – order in which enzyme isolated from strain of bacteria.
5. A linear DNA fragment and a plasmid has three restriction sites for EcoRIhow many fragments will be produced from linear DNA and plasmid respectively.
Ans.Number of fragments of linear DNA = 4
Number of fragments of plasmid = 3
6. An extra chromosomal segment of circular DNA of a bacterium is used to carry gene of interest into the host cell. What is the name given to it?
Ans. Plasmid.
7. Identify the recognition sites in the given sequences at which E.coli will be cut and make sticky ends.
5´-GAATTC-3´
3´-CTTAAG-5´
Ans. 5−G↓AATTC35−G↓AATTC3
3−CTTAA↑G53−CTTAA↑G5
8.Name the substance used as a medium in gel electrophoresis.
Ans.Agarose
9.What is Bioconversion?
Ans.Bioconversion refers to the process by which raw material are converted to specific product by microbial, plant or animal cell.
10.Name the bacterium that yields thermostable DNA polymerase.
Ans.Thermusaquaticus.
11.Which enzymes are known as “molecular Scissors”?
Ans.Restriction Endonuclease.
12.Name the commonly used vector for trans formation in plant cell?
Ans.Agrobacterium tumefacien.
13.Name the technique used for amplification of DNA?
Ans.Polymerase Chain Reaction.
14.Name the enzyme responsible for removal of 5 – phosphate group from nucleic acid?
Ans.Alkaline Phosphates.
15.Who isolated Restriction enzymes for the first time?
Ans.Warner Arber & Hamilton Smith.
16.Why do eukaryotic cells do not contain restriction enzymes?
Ans.Because in eukaryotic cell, DNA is heavily methylated.
17.Why does DNA moves towards anode in gel electrophoresis.
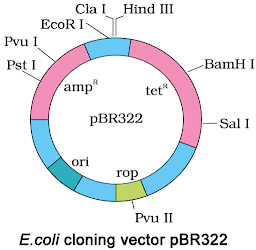
Ans.Because of presence of phosphate group, DNA is negatively charged &∴∴ moves towards anode.
2 Mark Questions
Chapter 11
Biotechnology Principles and Processes
2 Marks Questions
1.Name two main steps which are collectively referred to as down streaming process. Why is this process significant?
Ans.Separation and Purification
This process is essential because before reaching into market, the product has to be subjected for clinical trial and quality control.
2. How does plasmid differ from chromosomal DNA?
Ans.
| Plasmid DNA | Chromosomal DNA | ||
| (i) (ii) (iii) | Circular DNA Occurs only in bacterial cells Used as Vector in rDNA technology | (i) (ii) (iii) | Linear DNA Occurs in nucleus of eukaryotic cells and bacterial cell. Not used as vector in rDNA technology. |
3. A bacterial cell is shown in the figure given below. Label the part ‘A’ and ‘B’. Also mention the use of part ‘A’ in rDNA technology.
Ans. A- Plasmid, B – Nucleoid
Plasmid is used as vector to transfer the gene of interest in the host cell.
4. Mention two classes of restriction enzymes. Suggest their respective roles.
Ans.Exonucleases and endonucleases
- Exonucleases remove nucleotides from the ends of the DNA.
- Endonucleases cut DNA at specific sites beween the ends of DNA.
5. In the given process of separation and isolation of DNA fragments, someof the steps are missing, Complete the missing steps –
A : Digestion of DNA fragments using restriction endonucleases
↓
B : ……………………………………………………..
↓
C : Staining with ethidium bromide
↓
D :Visualisation in U.V. light
↓
E : …………………………………………………….
↓
F : Purification of DNA fragments.
Ans.B – Gel Electrophoresis
E – Elution
6.Write any two properties of restriction endonuclease enzymes?
Ans.(i) Each Restriction endonuclease functions by inspecting the length of DNA sequence & bindto DNA at the recognition Sequence.
(ii) It cuts the two strands of DNA at specific point in their sugar – phosphate backbone.
7.What are ‘Selectable marker’? What is their use in genetic engineering?
Ans.A selectable marker is a gene which helps in selecting those host cells which contains thevector& eliminating the non–transformanteg – gene encoding resistance to antibiotics are usefulSelectable markers as they allow Selective growth of transformants only.
8.How can the desired product formed after genetic engineering be produced on a commercial scale?
Ans.The product obtained from genetic engineering is subjected to a series of processes collectivelycalled downstream processing before it made into final processes involved in downstreamprocessing are :- Separation & purification.
9.What is “Insertional Inactivation”?
Ans.If a recombinant DNA is inserted within the coding Sequence of enzyme B–galactosidase. Thisresults into inactivation of enzyme which is referred to as “Insertional Inactivation”. The presenceof chromogenic Substrate gives blue–coloured colonies if the plasmid in bacteria does not have aninsert presence of insert results into insertional inactivation &the colonies do not produce anycolor.
10.What are the two basic techniques involved in modern Biotechnology?
Ans.The two basic techniques involved in modern Biotechnology are:-
a)Genetic Engineering is the technique of altering the nature of genetic material or introduction of it into another host organism to change its phenotype.
b)Techniques to facilitate the growth & multiplication of only the desired microbes or cells in large number under sterile conditions for manufacture
11.Represent diagrammatically the E. coli. Cloning vector ββ PBR 322.
Ans.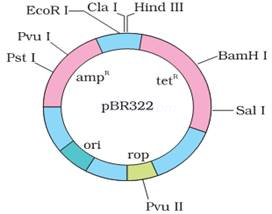
12.Differentiate between plasmid DNA and chromosomal DNA?
Ans.Plasmid DNA is extranuclear DNA, found in protoplasmic whereas chromosomal DNA is the nuclear or genetic DNA which is found within the nucleus.
13.What is the role of enzyme “Ligase” in genetic Engineering?
Ans.Enzyme “Ligase” acts as molecular Suture which helps in joining two pieces of DNA. The Joining process requires ATP as it derive energy to construct phosphodiester bond between cohesive ends.
14.Name the components a bioreactor must possess to achieve the desired product?
Ans.Enzyme “Ligase” acts as molecular Suture which helps in joining two pieces of DNA. The Joining process requires ATP as it derive energy to construct phosphodiester bond between cohesive ends.
15.The following proteins of given molecular weight are Subjected to Get electrophoresis. Write the order of Sequence in which these proteins are isolated in a gel?
| S.no. | Proteins | Mol.wt |
| 1. | Albumin | 23,000 |
| 2. | Keratin | 48,000 |
| 3. | Myosin | 1,25,000 |
| 4. | Haemoglobin | 84,000 |
| 5. | Ribozyme | 62,000 |
| 6. | Insulin | 1,14,000 |
Ans.The sequence of proteins obtained from top to bottom in a gel:-
Myosin > Insulin >Haemoglobin> Ribozyme > Keratin > Albumin.
16.How is gene Z used as a marker?
Ans.Lac Z gene codes for enzyme Β-galactosidase, if a recombinant DNA is inserted within thecoding sequence of an enzyme Β-galactosidase. This results into inactivation of enzyme. Thebacterial colonies whose plasmid does not have an insert produce blue colour but those with aninsert do not produce any colour.
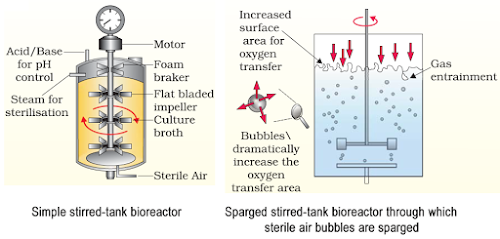
17.What is Bioreactor? What are the advantages of Stirred tank Bioreactor over Shake flask. Show diagrammatically a simple Stirred tank Bioreactor?
Ans.Bioreactors are large vessels in which raw materials are biologically converted into specificproteins using microbial, plant, animal or human cells. The advantages of Bioreactor over shakeflask are :-
a)It provides optimal conditions for achieving desired product by providing optimum growth conditions eg. temp, pH etc.
b)Small volume of cultures can be withdrawn periodically from bioreactor to test the sample.
c)It has an agitation system, temp control system, from control system & pH control system
d)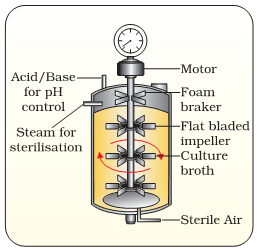
3 Mark Questions
Chapter 11
Biotechnology Principles and Processes
3 Marks Questions
1. Since DNA is a hydrophillicmoelcule, it cannot pass through cell membranes. Name and explain the technique with which the DNA is forced into (ii) a bacterial cell (ii) a plant cell (iii) an animal cell.
Ans. (i) Chemical treatment and exposure to cold and high temp.(42°C) alternatively. (Bacterial cell)
(ii) Biolistics or gene gun. (Plant cell)
(iii) Micro-injection. (animal cell) Explanation – Refer page 200, biology Text Book for class XII.
2. How will you obtain purified DNA from a cell?
Ans.Cells are treated with appropriate enzymes to release DNA. Lysozyme (bacteria), cellulase (plant cells), chitinase (fungus).
- RNA and proteins are removed by treatment with ribonuclease and protease enzymes respectively.
3. In recombinant DNA technology, vectors are used to transfer a gene of interest in the host cells. Mention any three features of vectors that are most suitable for this purpose.
Ans.(i) Have origin of replication(Ori)
(ii) Have a selectable marker
(iii) Have at least one recognition site.
4. Why is “Agrobacterium–mediated genetic engineering transformation” inplants considered as natural genetic engineering?
Ans.Agrobacterium tumefaciens is a pathogen in many dicot plants. It is able to deliver a piece of DNA (T–DNA) to transform normal plant cell into a tumor and directs these tumor cells to produce the chemicals required by pathogen.
5. Observe the given sequence of nitrogenous bases on a DNA fragment and answer the following question –
5´ – CAGAATTCTTA – 3´
3´ – GTCTTAAGAAT – 5´
(a) Name a restriction enzme which can recognise this DNA sequence.
(b) Write the sequence after digestion.
(c) Why are the ends generated after digestion called sticky ends?
Ans. (a) EcoRI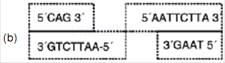
(c) These are named sticky ends, because they form hydrogen bonds with their complementary cut parts.
6. A selectable marker is used in the section of recombinants on the basis of their ability to produce colour in presence of chromogenic substrate.
(a) Mention the name of mechanism involved.
(b) Which enzyme is involved in production of colour?
(c) How is it advantageous over using antibiotic resistant gene as a selectable marker?
Ans. (a) Insertional inactivation
(b) b-galactosidase.
(c) Selection of recombinants due to inactivation of antibiotics requires simultaneous plating on two plates having different antibiotics. (Refer page 200 NCERT Biology for class XII).
7.Mention the important properties which a good vector must possess?
Ans.The important properties which a good vector must possess are :-
i) Size :- The vector must have small size so that it is easier to purify & isolate.
ii) Origin of replication :- This is a sequence of base pairs where replication starts. Any piece of DNA linked to this sequence can be made to replicate within its host cell & thus, controls the copy number of linked DNA.
iii) Selectable Marker :- A marker is a gene which helps in selecting those host cells which contain the vector & eliminating the non – transformants Common Selectable marker include gene encoding resistance to antibiotics.
iv) Cloning Sites :- The vector Should have a few or at least one unique recognition site to link the foreign / alien DNA. Presence of a particular recognition site enables the particular restriction enzyme to cut the vector.
8.Describe any three vectors less method of introducing the rDNA into a competent host cell?
Ans. i) Transformation :- In order to force bacteria to take up the plasmid, the bacterial cell must first be made competent to take up DNA. This is done by treating them with specific concentration of divalent cationeg. Ca2+ which increases the efficiency with which DNA enters the bacterium through pores in its cell wall Recombinant DNA can then be forced into such cells by incubating the cells with recombinant DNA on ice, followed by placing them at 420 C& then putting them back into ice. This enables the bacteria to take up the recombinant DNA.
ii) Microinjection :- recombinant DNA is directly injected into the nucleus of an animal cell using a micro – needle of tip with diameter (~ 4mm)
iii) Biolistics / Gene gun :- cells are bombarded with high velocity micro – particles of gold or tungsten coated with DNA.
9.Why is Agrobacterium mediated genetic transformation described as Natural Genetic engineering in plants?
Ans.Agrobacterium tumefacien, anatural pathogen of Several dicot plants is able to deliver a piece of DNA known as “t – DNA” to transform normal plant cell into a tumor & direct gene transfer transform tumor cells to produce chemicals required by pathogen . The tumor inducing (Ti) plasmid of
Agrobacterium tumefacien has now been modified into a cloning vector which is no more pathogenic to plant but is still able to use the mechanism to deliver genes of our interest into a variety of plants Since Agrobacterium tumefacien has the natural ability to donate a part of its DNA to the plant during infection. This property of Agrobacterium is exploited and a gene of interest is ligated into T-DNA so that it automatically gets transformed into plant cell thus, Agrobacterium tumefacien is known as “Natural Genetic Engineer” of plants.
10.Mention the important tools required for genetic engineering technology?
Ans.The process of genetic engineering is accomplished only when we have following key tools :-
a)Restriction enzymes:- Restriction enzymes are a group of endonucleases which cut the DNA at Specific position anywhere in its length. Each restriction endonuclease functions by inspecting the length of DNA & binds to DNA at the recognition Sequence.
b)Cloning Vector:- The DNA molecule which carry the desired DNA Segment of an organism & transfer it to cell or DNA of another organism is called cloning vector.
c)Desired foreign DNA:- The segment of DNA containing genes having desired characters & which are being transferred into genome of another cell with the help of vector is called foreign DNA.
5 Marks Questions
Chapter 11
Biotechnology Principles and Processes
5 Marks Questions
1. The development of bioreactors is required to produce large quantities of products.
(a) Give optimum growth conditions used in bioreactors.
(b) Draw a well labelled diagram of simple stirred – tank bioreactor.
(c) How does a simple stirred – tank’ bioreactor differ from sparged stirred – tank’ bioreactor?
Ans. (i) Temperature, pH, susbtrates, salts, vitamins and oxygen.
(ii) (a) simple stirred–tank bioreactor
(iii) The stirrer facilitates even mixing and oxygen availability throughout simple–stirred tank bioreactor, whereas in case of sparged stirred-tank bioreactor, air is bubbled throughout the reactor for proper mixing.
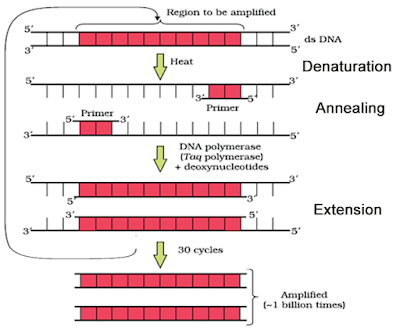
2. In the given figure, one cycle of polymerase chain reaction (PCR) is shown-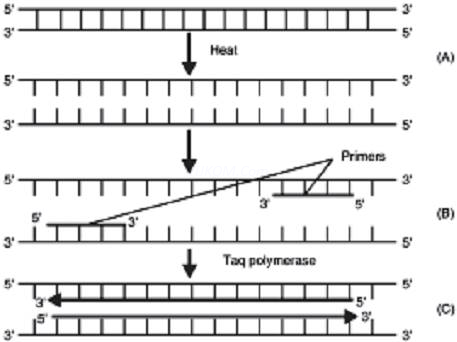
(a) Name the steps A, B and C.
(b) Give the purpose of each of these steps.
(c) State the contribution of bacteriumThermusaquaticus in this process.
Ans. (a) Denaturation – Heat denatures DNA to separate complementary strands.
(b) Annealing : Primers hybridises to the denatured DNA strands.
(c) Extension : Extension of primers resulting in synthesis of copies of target DNA sequence. Enzyme Tag polymerase is isolated from the bacterium Thermusaquaticus. This enzyme induces denaturation of double stranded DNA at high temperature.
3. Study the figure of vector pBR322 given below in which foreign DNA is ligated at the Bam H1 site of tetracyline resistance gene.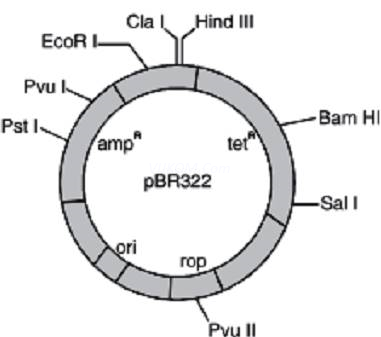
Answer the following questions :
(a) Mention the function of rop.
(b) What will be the selectable marker for this recombinant plasmid and why?
(c) Explain transformation.
Ans. (a) ‘rop’ codes for the proteins involved in the replication of plasmid
(b) Selectable marker – ampicillin resistance gene. It will help distinguishing trAns.formants from non-trAns.formants after plating them on ampicillin containing medium.
(c) Transformation – It is the phenomenon by which the DNA isolated from one type of cell and introduced into another type and is able to bring about some of the properties of former to the later.
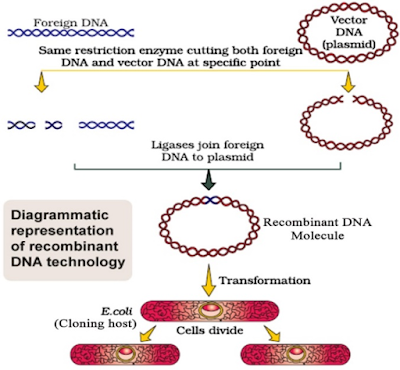
4.Describe the various steps involved in Recombinant DNA technology with the help of a well labeled. Diagram?
Ans. i) Identification of DNA with desirable Genes:- Other molecules in the target cell can beremovedby appropriate treatment & purified DNA ultimately precipitates out after addition ofchilled ethanol.
ii) Cutting the DNA at specific location :- After having cut the source DNA as well as vector DNA with Specific restriction enzyme, the cut out “gene of interest” from the source DNA & the cut vector with space are mixed & ligase is added.
iii)Insertion of Recombinant DNA into host cell :- Recipient cells after making them competent to receive takes up DNA in its surrounding. Recombinant DNA is introduced into suitable host cell by vector – based or vector – less method.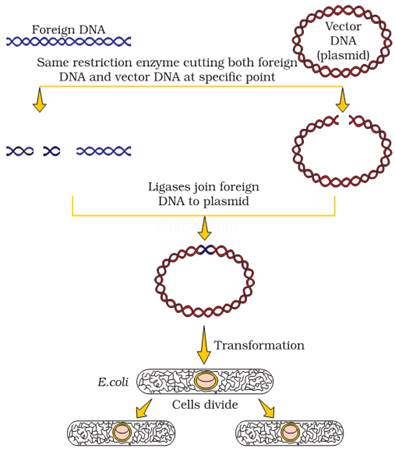
iv)Selection & Screening :- If a recombinant DNA bearing gene for resistance to an antibiotic is trAns.ferred into E-coli the host – cell become trAns.formed into ampicillin – resistant cells. Due to this amp gene one is able to select a trAns.formed cell in the presence of ampicillin. This amp r gene is called selectable marker.
v)Obtaining the foreign Gene product :- After having cloned the gene of interest & having optimized the conditions to induce expression of the target protein, one has to consider producing it on large scale.
5.Expand PCR? Describe the different Steps involved in this technique?
Ans. PCR stands for polymerase chain reaction.It is a technique for amplification of gene of interest
or to obtain multiple copies of DNA of interest.
The PCR requires primers, taq polymerase, target sequence, DNA sample &deoxyribonucleotides.
PCR includes number of cycles for amplifying DNA of interest invitro. Each cycle has three steps :-
a)DENATURATION:- The first step is denaturation of SNA sample in a reaction mixture to 940 c. During this step, DNA strand gets separated.
b)RENATURATION / ANNEALING:- The temperature is allowed to cool down to 500c to allow two oligo-nucleotide primers to anneal to complementary sequence in DNA molecule.
c)EXTENSION:- The temperature is raised to 750c. At this temperature, taq – polymerase initiates DNA Synthesis at 3-OH end of primer.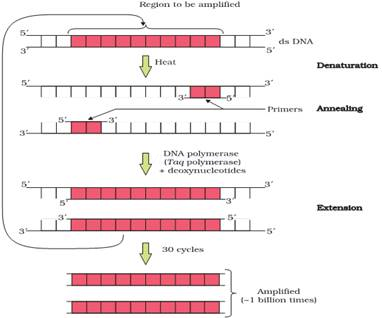
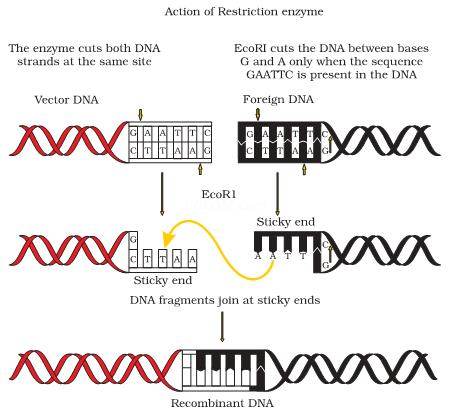
6.What are Restriction enzyme? Why do bacteria have these restriction enzymes. Show diagrammatically a restriction enzyme its recognition & the product it produces?
Ans. Restriction enzymes are endonucleases which recognize a specific sequence within DNA andcut the DNA within that sequence at a specific point. In bacteria, these restriction enzymes operatea modification restriction system which modifies & cuts the foreign DNA entering into the bacterial cell& thus, provides immunity to bacterial cell.
Name of Restriction enzyme- EcoRISubstrate DNA on which it acts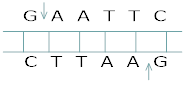
Biotechnology: Principles and Processes Class 12 Biology MCQs
1. Biolistics (gene gun) is suitable for
(a) introducing rDNA into plant cells
(b) introducing rDNA into animal cells
(c) disarming the pathogen vectors
(d) DNA fingerprinting.
Answer
Answer: a
2. In genetic engineering experiments, restriction enzymes are used for
(a) viral DNA
(b) bacterial DNA
(c) eukaryotic DNA
(d) any type of DNA.
Answer
Answer: b
3. The DNA fragments produced by the use of restriction endonucleases can be separated by
(a) polymerase chain reaction
(b) gel electrophoresis
(c) density gradient centrifugation
(d) any of the above.
Answer
Answer: b
4. Plasmids in bacterial cells are
(a) extra-chromosomal DNA, which cannot replicate
(b) extra-chromosomal DNA, which can . self-replicate
(c) extra DNA associated with the genome
(d) extra DNA, associated with the genome, but cannot replicate.
Answer
Answer: b
5. The DNA polymerase enzyme used in PCR is obtained from
(a) Thermus aquaticus
(b) Escherichia coli
(c) Agrobacterium tumefaciens
(d) Salmonella typhimurium.
Answer
Answer: a
6. While isolating DNA from bacteria, which of the following enzymes is not used? [NCERT Exemplar]
(a) Lysozyme
(b) Ribonuclease
(c) Deoxyribonuclease
(d) Protease
Answer
Answer: a
7. Significance of ‘heat shock’ method in bacterial transformation is to facilitate [NCERT Exemplar]
(a) binding of DNA to the cell wall
(b) uptake of DNA through membrane transport proteins
(c) uptake of DNA through transient pores in the bacterial cell wall
(d) expression of antibiotic resistance gene
Answer
Answer: c
8. Which of the following steps are catalysed by Taq polymerase in a PCR reaction? [NCERT Exemplar]
(a) Denaturation of template DNA
(b) Annealing of primers to template DNA
(c) Extension of primer end on the template DNA
(d) All of the above
Answer
Answer: c
9. Which of the given statement is correct in the context of observing DNA separated by agarose gel electrophoresis? [NCERT Exemplar]
(a) DNA can be seen in visible light
(b) DNA can be seen without staining in visible light
(c) Ethidium bromide stained DNA can be seen in visible light
(d) Ethidium bromide stained DNA can be seen under exposure to UV light
Answer
Answer: d
10. ‘Restriction’ in Restriction enzyme refers to [NCERT Exemplar]
(a) cleaving ofphosphodiester bond in DNA by the enzyme
(b) cutting of DNA at specific position only
(c) prevention of the multiplication of bacteriophage in bacteria
(d) all of the above
Answer
Answer: c
11. The role of DNA ligase in the construction of a recombinant DNA molecule is [NCERT Exemplar]
(a) formation of phosphodiester bond between two DNA fragments
(b) formation of hydrogen bonds between sticky ends of DNA fragments
(c) ligation of all purine and pyrimidine bases
(d) none of the above
Answer
Answer: b
12. In an experiment, recombinant DNA bearing ampicillin-resistance gene is transferred into E.coli cells. The host cells are then cultured on a medium containing ampicillin. The result will be
(a) both transformants and non-transformants cannot survive.
(b) both transformants and non-transformants can survive.
(c) transformants only and not the non-transformants can survive.
(d) transformants cannot survive, but non-transformants can not.
Answer
Answer: c
13. The _______ in a vector helps in identifying the transformants and eliminating the non-transformants.
Answer/Explanation
Answer:
Explaination: Selectable marker.
14. When the enzyme _______ is inactivated in E.coli, the transformants do not produce any colour in the presence of a chromogenic substrate.
Answer/Explanation
Answer:
Explaination: β-galactosidase.
15. _______ is the method of bombarding high velocity microparticles of gold or tungsten coated with DNA, into plant cells.
Answer/Explanation
Answer:
Explaination: Biolistics/gene gun.
16. To isolate DNA from fungal cells for biotechnology experiments, enzyme _______ is necessary.
Answer/Explanation
Answer:
Explaination: Chitinase.
17. _______ is used as cloning vector for transformation in plant cells.
Answer/Explanation
Answer:
Explaination: Agmbacterium.
18. Downstream processing involves separation and _______ .
Answer/Explanation
Answer:
Explaination: Purification.
19. _______ is the process of extraction of DNA from the separated bands of DNA in agarose gel.
Answer/Explanation
Answer:
Explaination: Elution.
20. _______ gene in the E.coli vector, pBR 322 codes for enzymes/proteins involved in the replication of the plasmid.
Answer/Explanation
Answer:
Explaination: Rop.
21. _______ are the E.coli enzymes that remove the nucleotides from the ends of DNA strands.
Answer/Explanation
Answer:
Explaination: Exonucleases.
22. _______ is an autonomously replicating, circular, extra-chromosomal DNA in bacterial cells.
Answer/Explanation
Answer:
Explaination: Plasmid.
23. Match the items in Column I with those in Column II.
| Column I | Column II |
| A. Sea weeds | 1. Gel electrophoresis |
| B. Staining of DNA | 2. Source of Agarose |
| C. Separation of DNA fragments | 3. Isolation of DNA from the gel |
| D. Elution | 4. Ethidium bromide |
Answer/Explanation
Answer:
Explaination: A – 2, B – 4, C – 1, D – 3
24. Match the Column l with the Column II.
| Column I | Column II |
| A. Competent host | 1. Separation and purification |
| B. Cloning vector | 2. Culturing of cells in large volumes |
| C. Downstream processing | 3. Taq polymerase |
| D. PCR | 4. Divalent cation (Ca2+) |
| E. Bioreactor | 5. pBR 322 |
| 6. Gel electrophoresis |
Answer/Explanation
Answer:
Explaination: A – 4, B – 5, C – 1, D – 3, E – 2
25. Since DNA fragments are positively charged they move towards the anode. [True/False]
Answer/Explanation
Answer:
Explaination: False.
26. Agarose, the most commonly used matrix in gel electrophoresis is obtained from fungi. [True/False]
Answer/Explanation
Answer:
Explaination: False.
27. The DNA fragments separate according to their size through agarose gel during electrophoresis. [True/False]
Answer/Explanation
Answer:
Explaination: True.
28. The cloning vector pBR 322 has three antibiotic-resistance genes. [True/False]
Answer/Explanation
Answer:
Explaination: False.
29. The vector DNA and foreign DNA are cut by the same restriction endonuclease. [True/False]
Answer/Explanation
Answer:
Explaination: True.
Directions (Q30 to Q32): Mark the odd one in each of the following groups.
30. Cellulose, Lysozyme, Chitinase, Endonuclease
Answer/Explanation
Answer:
Explaination: Endonuclease.
31. Hind III, EcoRI, Sal I, Rop
Answer/Explanation
Answer:
Explaination: Rop.
32. Denaturation, Elution, Annealing, Extension
Answer/Explanation
Answer:
Explaination: Elution.
33. How has European Federation of Biotechnology (EFB) defined biotechnology?
Answer/Explanation
Answer:
Explaination: The European Federation of Biotechnology defines biotechnology as the integration of natural science and organisms, cells or parts thereof and molecular analogues, for products and services; it encompasses both .traditional and modem molecular biology.
34. Name the technique that is used to alter the chemistry of genetic material (DNA/RNA) to obtain the desired result. [Delhi 2016C]
Answer/Explanation
Answer:
Explaination: Genetic engineering.
35. What is meant by gene cloning?
Answer/Explanation
Answer:
Explaination: Gene cloning refers to the process in which a numer of identical copies of a gene of interest are made by introducing the gene into an appropriate host using a suitable Vector.
36. Why is it not possible for an alien DNA to become part of a chromosome anywhere along its length and replicate normally? [AI 2014]
Answer/Explanation
Answer:
Explaination: The alien DNA has to be linked to a specific sequence of DNA, called origin of replication, on the chromosome; this origin of replication is responsible for initiation of replication.
37. Name the specific sequence of DNA in a plasmid that the ‘gene of interest’ ligates with, to enable it to replicate. [AI 2013C]
Answer/Explanation
Answer:
Explaination: Origin of replication (Ori)
38. Write the two components of the first artificial recombinant DNA molecule constructed by Cohen and Boyer. [Foreign 2014]
Answer/Explanation
Answer:
Explaination: An antibiotic-resistance gene and the plasmid of Salmonella typhimurium.
39. Name the two enzymes that are essential for constructing a recombinant DNA. [Delhi 2017C]
Answer/Explanation
Answer:
Explaination: Restriction endonuclease and DNA ligase.
40. In the year 1963, two enzymes responsible for restricting the growth of bacteriophage in E.coli were isolated. How did the enzymes act to restrict the growth of the bacteriophage? [AI 2011C]
Answer/Explanation
Answer:
Explaination:
– One of them (exonucleases) added methyl groups to DNA.
– The other (endonucleases) cut the DNA at specific points within the DNA.
41.Name the enzyme that helps to join DNA fragments. [AI 2017C]
Answer/Explanation
Answer:
Explaination: DNA ligase
42. Mention the role of Restriction enzyme in Recombinant DNA technology. [Delhi 2017C]
Answer/Explanation
Answer:
Explaination: They act as molecular scissors to cut DNA at specific locations.
43. What does ‘R’ stand for, in the restriction endonuclease, EcoRI?
Answer/Explanation
Answer:
Explaination: R stands for RY 13, the strain of E.coli bacterium.
44. A plasmid and a DNA sequence in a cell need to be cut for producing recombinant DNA. Name the enzyme that acts as molecular scissors to cut the DNAs.
Answer/Explanation
Answer:
Explaination: Restriction endonucleases.
45. Suggest a technique to a researcher who needs to separate fragments of DNA. [Delhi 2016]
Answer/Explanation
Answer:
Explaination: Gel electrophoresis.
46. Name the material used as matrix in gel electrophoresis and mention its role. [AI 2014C]
Answer/Explanation
Answer:
Explaination: Agarose; it provides the sieving effect for the DNA to resolve according to their size.
47. Why do DNA-fragments move towards the anode during gel electrophoresis? [CBSE 2018C, Delhi 2011C; HOTS]
Answer/Explanation
Answer:
Explaination: DNA-fragments are negatively charged and hence, move towards the anode.
48. What is the role of ethidium bromide during agarose-gel electrophoresis of DNA fragments?
Answer/Explanation
Answer:
Explaination: The gel is stained by ethidium bromide, to view the separated DNA bands when exposed to UV light.
49. Which main technique and instrument is used to isolate DNA from a plant cell? [CBSE Sample Paper 2013, 2014]
Answer/Explanation
Answer:
Explaination:
– Centrifugation is the technique.
– Centrifuge is the instrument.
50. Name two commonly used vectors for genetic engineering.
Answer/Explanation
Answer:
Explaination: Plasmids and bacteriophages.
51. Why are engineered vectors preferred these days?
Answer/Explanation
Answer:
Explaination:
(i) Engineered vectors facilitate easy linking of foreign DNA.
(ii) They help in selection of recombinants.
52. Mention the uses of cloning vectors in biotechnology. [Delhi 2011]
Answer/Explanation
Answer:
Explaination:
(i) Cloning vectors are used to make multiple copies of the desired DNA/ gene.
(ii) They are used to transfer the gene of interest to the host cell.
53. Why is ‘plasmid’ an important tool in biotechnology experiments? [AI2013C]
Answer/Explanation
Answer:
Explaination: Since plasmids can replicate within the bacterial cell independently of the genomic DNA, any alien DNA ligated to it will also multiply, i.e. it is used as a vector as well as in gene cloning.
54. Why is it essential to have a ‘selectable marker’ in a cloning vector? [AI 2010; HOTS]
Answer/Explanation
Answer:
Explaination: It helps in identifying the recombinants from the non-recombinants.
55. Why are antibiotic-resistance genes used as markers in E.coli? [HOTS]
Answer/Explanation
Answer:
Explaination: It is because the normal E. coli cells do not have resistance to any such antibiotics.
56. Name two antibiotic-resistance genes in the pBR 322 of E.coli plasmid.
Answer/Explanation
Answer:
Explaination: Ampicillin-resistance gene and tetracyclin- resistance gene.
57. A plasmid without a selectable marker was chosen as vector for cloning a gene. How does this affect the experiment? [HOTS]
Answer/Explanation
Answer:
Explaination: In the absence of a selectable marker, it will not be possible to identify the recombinants from the non-recombinants.
58. State what happens when an alien gene is ligated at Sal I site of pBR 322 plasmid. [Delhi 2013C]
Answer/Explanation
Answer:
Explaination: The transformant loses the tetracycline- resistance.
59. State what happens when an alien gene is ligated at Pvu I site of pBR 322 plasmid. [AI 2013C]
Answer/Explanation
Answer:
Explaination: The transformant loses ampicillin-resistance.
60. How is Agrobacterium tumefaciens able to transform a normal plant cell into a tumour? [Delhi 2013C]
Or
Biotechnologists refer to Agrobacterium tumefaciens as a natural genetic engineer of plants. Give reasons to support the statement. [AI 2011]
Answer/Explanation
Answer:
Explaination: When it infects a plant cell, it delivers a part of its DNA, called ‘T-DNA’ (or Tumour- inducing (Ti) plasmid) into the plant cell and transforms it into a tumour cell.
61. How can retroviruses be used efficiently in biotechnology experiments in spite of them being disease causing? [AI 2013C]
Answer/Explanation
Answer:
Explaination: Retroviruses are disarmed (diseases-causing gene is removed/inactivated) and used as vectors to deliver the recombinant/alien DNA into animal cells.
62. Name the commonly used vector for transformation in plant cells.
Answer/Explanation
Answer:
Explaination: Agrobacterium tumefaciens.
63. How does an alien DNA gain entry into a plant cell by ‘biolistic’ method? [Foreign 2013]
Answer/Explanation
Answer:
Explaination: The cells are bombarded with high velocity micro-particles of gold or tungsten, coated with the alien DNA.
64. Mention the type of host cells suitable for the gene guns to introduce an alien DNA. [Delhi 2014]
Answer/Explanation
Answer:
Explaination: Plant cells.
65. Name the host cells in which micro-injection technique is used to introduce an alien DNA. [Foreign 2014]
Answer/Explanation
Answer:
Explaination: Animal cells.
66. Why is the enzyme cellulase needed for isolating genetic material from plant cells and not from animal cells? [Delhi 2013, 2010; HOTS]
Answer/Explanation
Answer:
Explaination: Cellulase is used to digest the cell wall (cellulose) of plant cells; animal cells have no cell wall and hence, it is not needed.
67. How can bacterial DNA be released from the bacterial cell for biotechnology experiments? [Delhi 2011]
Answer/Explanation
Answer:
Explaination: The bacterial cell has to be treated with lysozyme, to digest the cell wall to release DNA.
68. To use the restriction enzyme to cut the DNA, the DNA must be in a pine form, i.e. free from proteins and RNAs associated with it. How is it achieved?
Answer/Explanation
Answer:
Explaination:
– Proteins are removed by using proteases.
– RNAs are removed by using ribonucleases (RNases).
69. Write the names of the enzymes that are used for isolation of DNA from bacterial and fungal cells, respectively for Recombinant DNA technology. [AI, Foreign 2014]
Answer/Explanation
Answer:
Explaination:
– Lysozymes for bacterial cells.
– Chitinase for fungal cells.
70. Write the importance of the bacterium Thermus aquaticus in polymerase chain reaction. [Delhi 2013C]
Answer/Explanation
Answer:
Explaination: It is the source of thermostable DNA- polymerase enzyme.