Question
Human insulin can be synthesised in a laboratory strain of Escherichia coli using recombinant DNA (rDNA) technology.
The starting point for the process is mRNA coding for insulin, isolated from human pancreas cells.
Four enzymes are needed:
- reverse transcriptase
- DNA polymerase
- restriction enzyme
- DNA ligase.
(a) (i) State the role of each of these enzymes in producing rDNA carrying the gene for human insulin.[4]
reverse transcriptase
DNA polymerase
restriction enzyme
DNA ligase
(ii) Outline the role of insulin in a healthy human.[3]
(iii) Describe and explain one advantage of treating diabetics with human insulin produced by rDNA technology.[2]
(b) It is possible to use rDNA technology to produce insulin with a slightly different structure from that of human insulin. The effect of the changed structure can then be investigated.
The activities of equal quantities of two insulins, both produced by E. coli, were compared in healthy, non-diabetic subjects:
- human insulin
- insulin X, in which the positions of two amino acids, lysine and proline, were exchanged. Lysine has a hydrophilic R group and proline has a hydrophobic R group.
The results of the investigation are shown in Fig. 3.1.
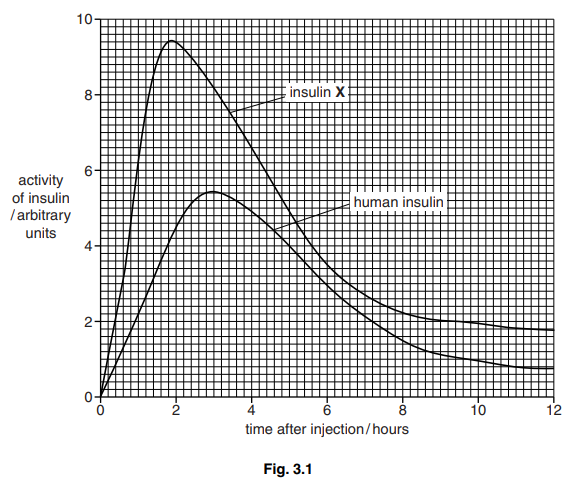
(i) With reference to Fig. 3.1 describe the differences in activity between human insulin and insulin X.[4]
(ii) Suggest how exchanging the position of two amino acids in the insulin molecule can result in differences in activity.[2] [Total: 15]
Answer/Explanation
Ans:
3 (a) (i) reverse transcriptase: produces (c)DNA from mRNA ;
DNA polymerase: produces double stranded DNA from, single stranded (DNA)/ cDNA ;
restriction enzyme: cuts, DNA/plasmid ;
DNA ligase: joins (gaps in) the sugar-phosphate backbone (of DNA) ;
(ii) 1 causes blood glucose concentration, to decrease/return to normal (from high) ;
2 (target cells are) liver/muscle ;
3 increased, absorption of glucose (from blood)/permeability of cell surface membrane to glucose ;
4 increased (rate of) respiration of glucose ;
5 idea of increased conversion of glucose to glycogen ;
6 inhibits secretion of glucagon/decreased gluconeogenesis ;
(ii) 1 identical to that produced by body ;
2 activity the same/fast response/no immune response ;
3 no need for animal insulin/AW ;
4 for religious reasons/for ethical reasons/for e.g. vegetarian ;
5 uncontaminated/pure ;
6 so no risk of disease ;
7 production very efficient/always available ;
8 extraction from animals, costly/ complex/limited by supply of animals ;
(b) (i) insulin X ora throughout for human insulin
1 greater initial increase in activity/AW ;
2 time of maximum activity/peak, earlier ; [1.9h v. 3h]
3 maximum activity/peak, greater ; [9.4 v 5.4 (a.u.)]
4 rate of decrease greater ;
5 activity always higher ;
6 comparative figures ; [see above]
(ii) 1 changes, tertiary/3D structure ;
2 affects binding to receptor (on cell surface membrane) ;
3 (this) affects production of second messenger ;
4 hydrophilic/hydrophobic, bonds different ;
5 AVP ; e.g. may affect, solubility in blood/transport in blood/rate at which broken down
Question
When preparing infertile women for in-vitro fertilisation (IVF), it is necessary to stimulate the growth and maturation of several ovarian follicles. This is done by giving daily injections of the glycoprotein hormone, follicle stimulating hormone (FSH).
Each molecule of FSH has quaternary structure and consists of two different polypeptide chains, α and β.
(a) Explain what is meant by quaternary structure.[1]
(b) Human FSH can be extracted from women’s urine (u-hFSH). A procedure involving the use of monoclonal antibodies is used to produce purified u-hFSH.
Suggest how monoclonal antibodies can be used to obtain purified u-hFSH from urine.[3]
(c) Recombinant human FSH (r-hFSH) can be produced by adding the genes coding for the α and β polypeptide chains of FSH to mammalian ovary cells.
Suggest why mammalian cells are needed to produce r-hFSH, rather than bacterial cells.[1]
(d) In IVF treatment, a second hormone, human chorionic gonadotrophin (hCG) is injected when mature ovarian follicles (Graafian follicles) have developed.
Draw a labelled diagram to show the structure of a mature ovarian follicle.[3]
(e) The effectiveness of r-hFSH was compared with that of u-hFSH. Women starting IVF treatment were randomly divided into two groups and given either r-hFSH or u-hFSH.
The differences between the two groups of women after FSH treatment are shown in Table 2.1.
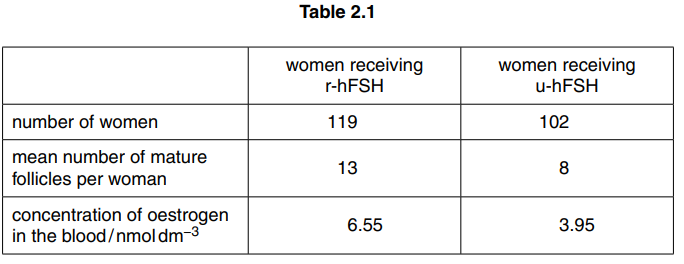
(i) With reference to Table 2.1, compare the effects of treatment with r-hFSH and u-hFSH and suggest explanations for the differences.[4]
(ii) The probability of the results for the mean number of mature follicles per woman occurring by chance is 0.002.
Explain what is meant by this probability.[2] [Total: 14]
Answer/Explanation
Ans:
2 (a) has more than one polypeptide ; A FSH has 2/α and β, polypeptides R has four has, prosthetic group/ non-protein part/ carbohydrate/ sugar ;
(b) 1 produce/ make, monoclonal antibodies specific to (u-h)FSH/ anti(u-h)FSH monoclonal antibodies ;
2 ref. to column/ framework, for, attachment/ immobilisation ; R test strip
3 urine, added to/ flows past/passed over, antibodies ;
4 (so) allowing, hormone/(h)FSH, to bind (to monoclonal antibodies) ;
5 treatment needed to release, hormone/(h)FSH (from monoclonal antibodies) ; I filtering
(c) 1 sugars need to be added/ glycosylation ; A bacteria cannot modify protein
2 needs, Golgi body /rough endoplasmic reticulum ; A bacteria lack, Golgi/rough endoplasmic reticulum
3 ref. to problems in bacteria with, introns /wrong promoter/ secretion/ora ;
(d) labels to correct recognisable structures
(secondary) oocyte ; R ovum
zona pellucida ;
corona radiata/ cumulus oophorus ;
fluid-(filled space)/ antrum ;
granulosa/follicle/ follicular, cells ;
theca ;
(e) (i) comparison
1 more mature follicles with r-hFSH ; ora
2 oestrogen (concentration), higher with r-hFSH ; ora
3 comparative data quote ; e.g. 13 v 8 mature follicles
OR 6.55 v 3.95nmol dm–3 oestrogen concentration
OR manipulated figures
e.g. difference of 5/ 2.6nmol dm–3 /
62.5% increase (r) follicles / 65.8% (r) oestrogen
explanation
4 (because) r-hFSH, purer/ more concentrated/ora
OR
(some) u-hFSH, damaged by extraction technique/ degraded ;
(ii) 1 difference/difference described, is significant ;
2 not due to chance ; A due to something other than chance
3 smaller than, critical value/ value for significance of, 0.05/ 5% ;
Question
(a) Describe how the gene coding for human insulin can be obtained and inserted into a plasmid vector. [8]
(b) Explain how bacteria can be genetically modified and then identified using antibiotic resistance genes. [7] [Total: 15]
Answer/Explanation
Ans:
9 (a) 1 obtain mRNA from β cells (of islets of Langerhans of pancreas) ;
2 reverse transcriptase ;
3 make (single-stranded) cDNA ;
4 DNA polymerase used to make cDNA double stranded ;
5 sticky ends created ; A description
6 (obtain) plasmids ;
7 cut with restriction, endonuclease/ enzyme ; A named e.g. EcoR1
8 ref. complementary sticky ends ;
9 cDNA/ insulin gene, mixed with plasmid ;
10 DNA ligase ;
11 seals nicks in sugar-phosphate backbone ; R anneals
(b) 1 (recombinant) plasmids mixed with bacteria ;
2 (some) bacteria, take up plasmids / transformed ;
3 heat shock / calcium chloride solution/Ca 2+ ions /electroporation ;
to identify bacteria containing plasmids
4 grow on, agar/medium, containing antibiotic (A) ; A ampicillin
5 plasmid contains, antibiotic (A)/ ampicillin, resistance gene(s) ;
6 bacteria with plasmid survive ; ora
to identify recombinant bacteria
7 replica plate ; A description e.g. sponge/ velvet pad/absorbent paper
8 (onto) agar/ medium, containing second antibiotic (B) ; A tetracycline
9 (tetR /B/ 2nd) resistance gene inactivated (by insertion of new, DNA/ gene)/AW ;
10 (ID) colonies from, 1st / ampicillin, plate that do not grow on, 2nd / tetracycline, plate ;
