- IB DP Biology 2025 SL- IB Style Practice Questions with Answer-Topic Wise-Paper 1
- IB DP Biology 2025 HL- IB Style Practice Questions with Answer-Topic Wise-Paper 1
- IB DP Biology 2025 SL- IB Style Practice Questions with Answer-Topic Wise-Paper 2
- IB DP Biology 2025 HL- IB Style Practice Questions with Answer-Topic Wise-Paper 2
D1.1 DNA replication
Why DNA replication?
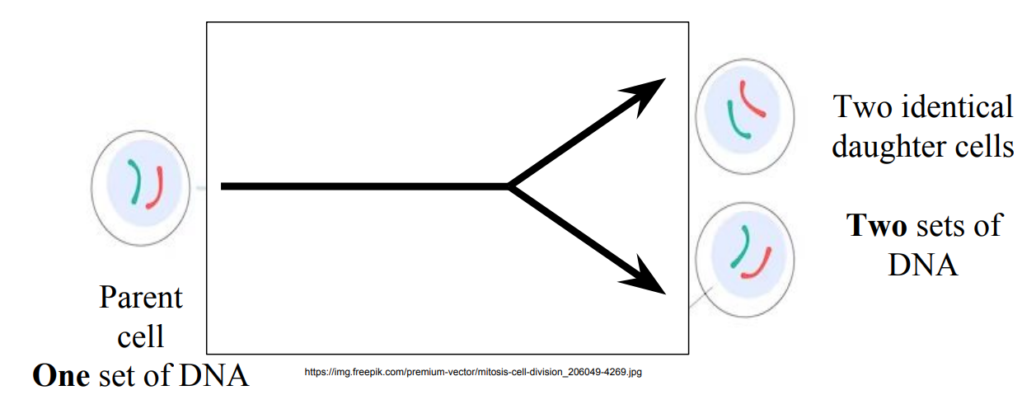
Process = Cell division
In cell division
One parent cell that divides will produce
Two genetically identical Daughter cells
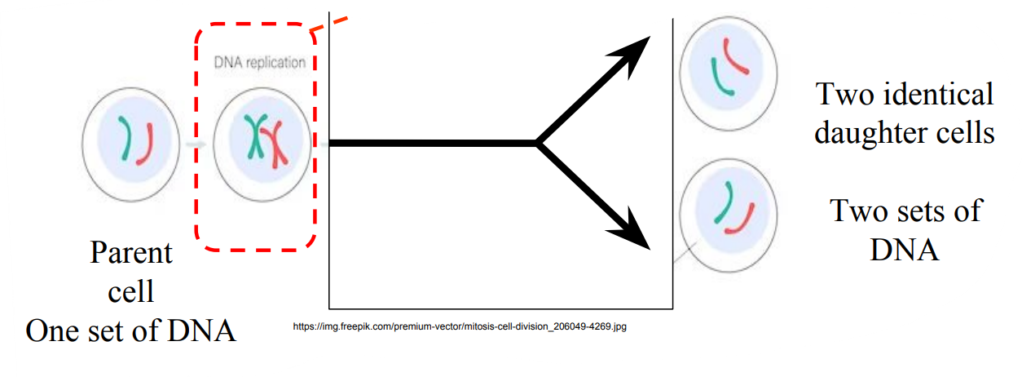
DNA replication needed first DNA replication produces exact copies
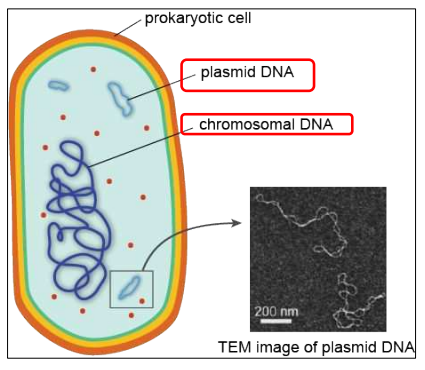
In Prokaryotes, DNA is present in the cytoplasm
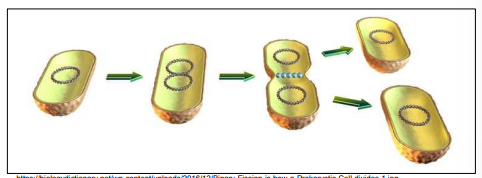
Binary fission in bacteria
Budding in yeast

In prokaryotes and unicellular eukaryotes, DNA replication and Cell division needed for Reproduction
In Eukaryotes, DNA is present in the nucleus, mitochondria and chloroplasts

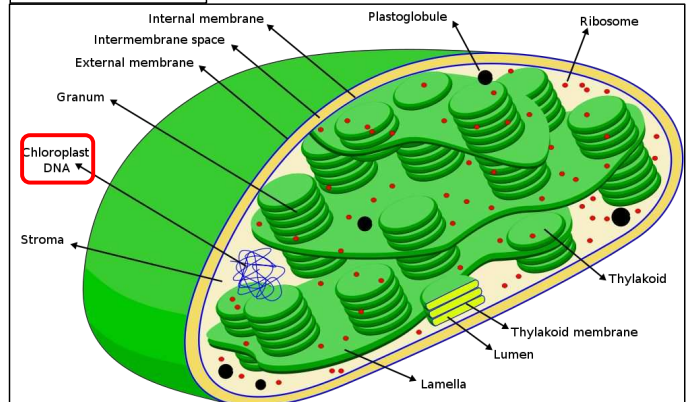
Chloroplast
Mitochondrion

In multicellular eukaryotes, DNA replication and Cell division needed for
- growth

- replacement of dead cells
- tissue repair
Complementary base pairing and semi-conservative DNA replication
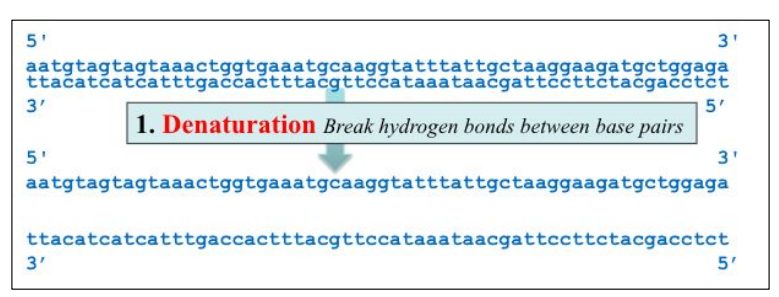
Denaturation
Separate the two strands
Breaking hydrogen bonds between pairs of bases
New strand ssDNA
Parent molecule ssDNA

Parent molecule dsDNA*
* dsDNA = double-stranded DNA
* ssDNA = single-stranded DNA
Synthesis of two new strands
Addition of new DNA nucleotides

Parent molecule dsDNA*
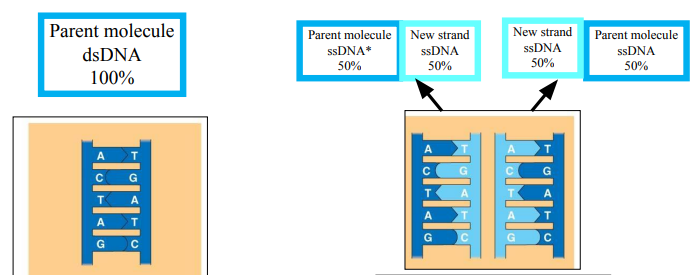
The daughter DNA contains
50% of the parent molecule
50% of the new molecule
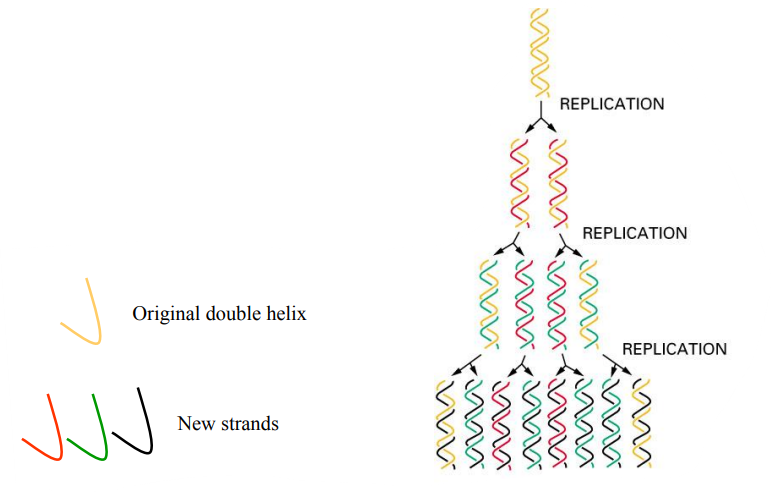
DNA replication is “semi-conservative”
Synthesis of new strands by complementing bases on both parent strands A/T and G/C base pairs
– Sequence of parent double helix = sequence of both daughters double helix
– DNA replication is highly accurate
– DNA sequence is conserved
“Semi-conservative”: KEEP 50%
After each DNA Replication,
Each double helix contains
One strand from the parent molecule
One newly made strand
Template strand
New strand


DNA replication and associated enzymes

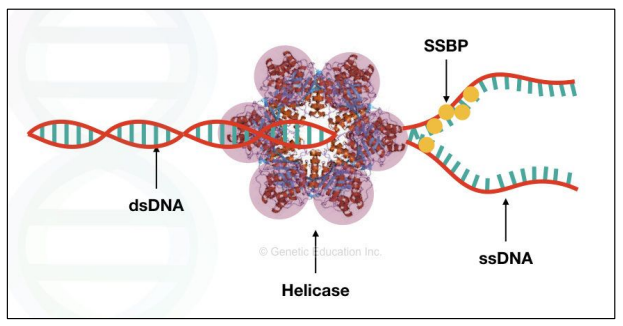
DNA Helicase = enzyme that unwinds the double helix and breaks hydrogen bonds between base pairs
Between A and T
Between G and C

DNA Polymerase = enzyme that complement both strands
Complements… A C G T
With… T G C A
Polymerase chain reaction and gel electrophoresis
Polymerase chain reaction
Polymerase Chain Reaction = PCR
Using DNA polymerase in a series of reactions in chain
to amplify a precise fragment of dsDNA = target DNA
Why use PCR?
– To detect the presence of a specific fragment in a sample
e.g. Is this plant transgenic?
– To amplify the coding sequence of a gene
e.g. Get the normal gene to be used in gene therapy
– To make DNA profiles
e.g. Is this my child/father/mother/ancestor?
Does the suspect’s DNA match the DNA found on the crime scene?
Number of repeats of a VNTR in organisms?
Organisms are closely/distantly related?



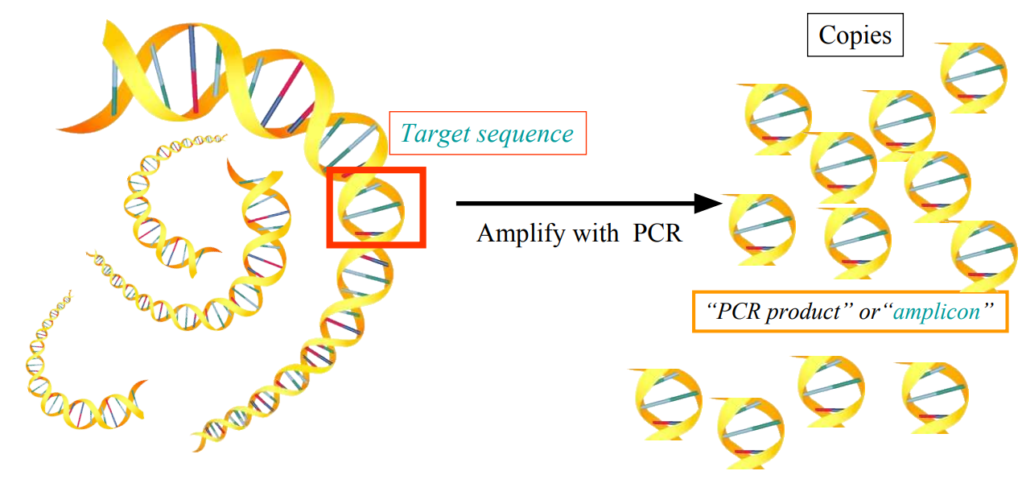
How to make sure only the target sequence will be copied?
Primers to focus the amplification on the target sequence only
Primers are short DNA sequences
They flank the target sequence
Only the target sequence will be amplified

One cycle of PCR

1. Denaturation
Breaking hydrogen bonds between paired bases
DNA Helicase not needed (and expensive)
Use heat = 95 degree C
2. Annealing
Binding of both primers to template DNA
Making new hydrogen bonds
Do not keep at 95 degree C
Cool down to 45-60 degree C
3. Extension
DNA Polymerase elongates both primers
Have to use the enzyme’s optimum temperature
Depends on source of enzyme
Usually use Taq DNA Polymerase
Use 72 degree C
Multiple cycles of PCR

Taq DNA Polymerase

Temperature used for denaturation = 95 degree C
– If use of human DNA Polymerase,
Enzyme denatured together with template DNA
– Need for an enzyme that can withstand 95 degree C without being denatured
Organisms that survive at high temperatures do possess such enzymes
Live bacteria found in hotsprings
i.e. Thermus aquaticus = Taq
Taq’s gene for DNA polymerase cloned into another bacteria
To produce (and sell) the enzyme Taq DNA Polymerase
Taq DNA Polymerase’s optimum temperature = 72 degree C
– Extension done at 72 degree C
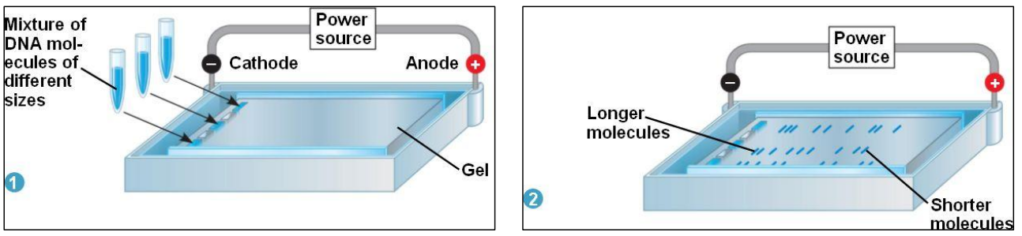
Gel electrophoresis
Goal = separate DNA fragments of different sizes (lengths)
How = DNA is negatively charged (Phosphate group)
– Electric current applied : DNA attracted to positive electrode = anode
Small fragments migrate faster than big fragments
– Small fragments will have migrated further away from origin than big fragments
Gel structure = sieve
Smaller molecules move faster
Bigger molecules move slower
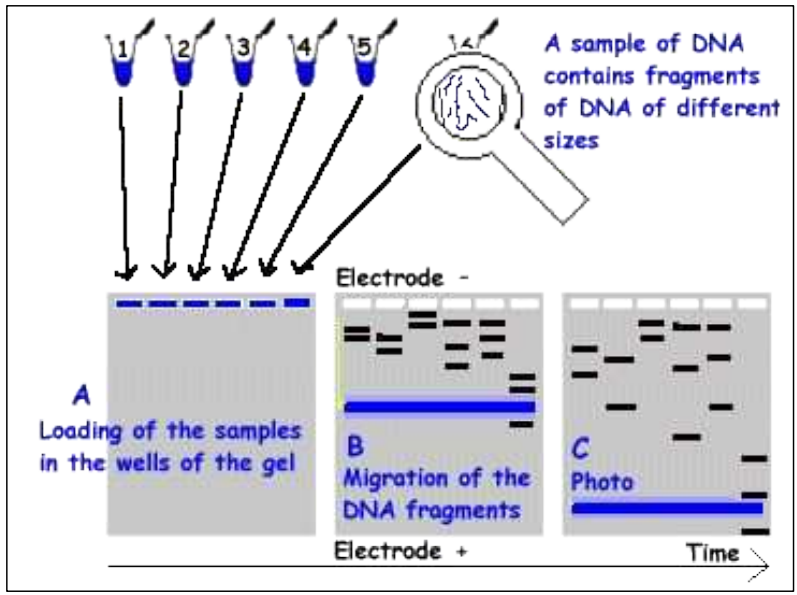
Estimating the size of DNA fragments

Uses in DNA profiling
DNA profiling
1. Making DNA profiles of organisms/DNA samples
2. Comparing these DNA profiles
3. Concluding about the identity(ies)
Same organism (or not)

Forensics
Who is the criminal?
Crime scene = Bands from criminal
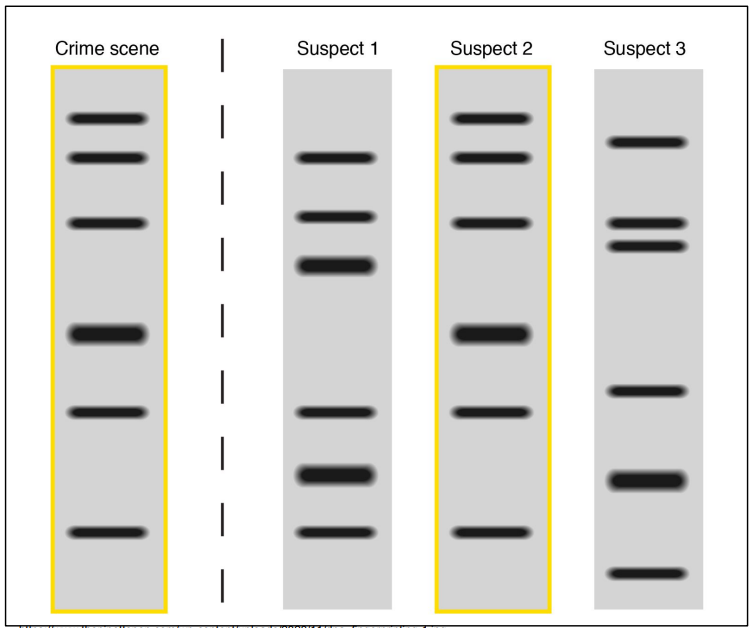
Only Suspect 2 has all crime scene’s bands
Therefore, Suspect 2 is the criminal
Paternity
Who is the father?
Child = Bands from mother + bands from father
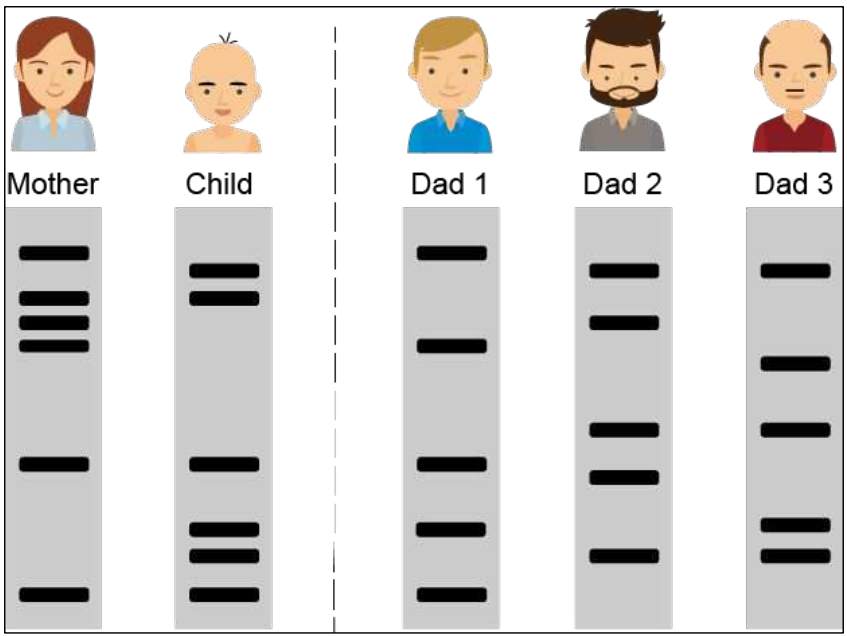
Bands that may be from mother
Bands that are from father

Only Dad 3 has all father’s bands
So, Dad 3 is the father
Directionality of DNA polymerases (HL only)

Template strand
New strand
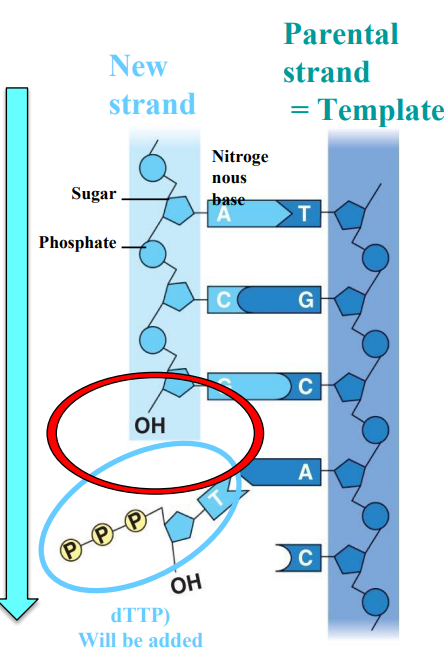
DNA polymerases have a directionality
From 5’ to 3’ of the new strand
A → T
T → A
C → G
G → C

Replication of one DNA strand “manually”

DNA Polymerase
Binds adjacent new DNA nucleotides with phosphodiester bonds

Leading strand and lagging strand (HL only)
Template strand
New strand
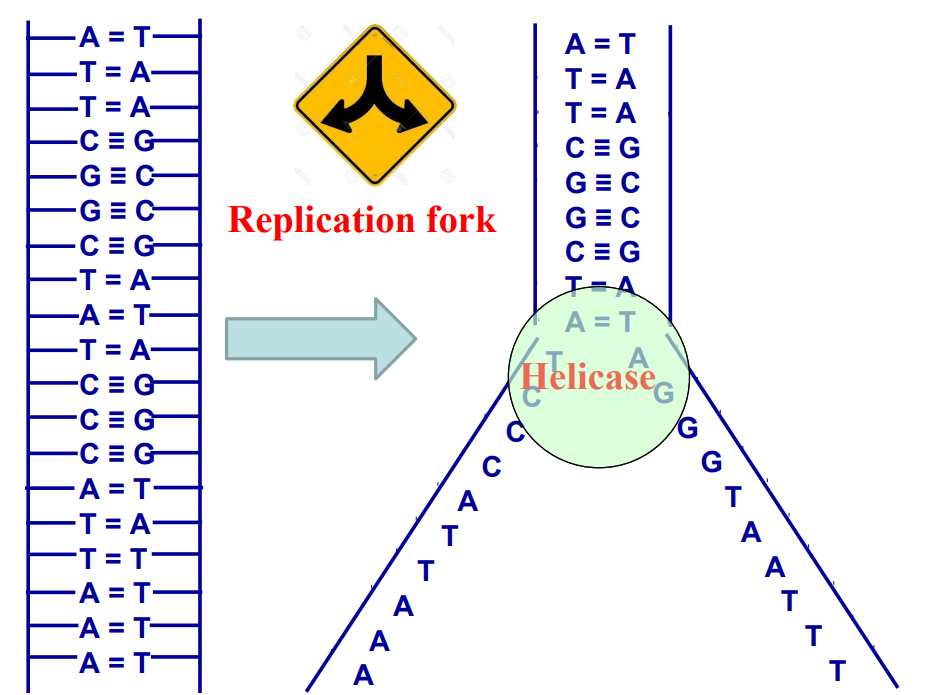
The DNA double helix has to be “opened/unwound”
So that proteins can access the information of both strands
To know which dNTPs have to be added, and in the right order
The replication starts at a Replication fork: DNA Helicase


Leading/Continuous strand

Lagging/Discontinuous strand



DNA replication and more associated enzymes (HL only)

DNA proofreading (HL only)

DNA Polymerase III can make mistakes
Wrong nucleotide added in 3’ of new strand
This could lead to harmful mutations
To avoid this,
DNA Polymerase III has an extra property = 3’ to 5’ exonuclease
Gets rid of mistakes = Proof-reading activity