| Biotechnology – Principles and Processes Refresher Course |
| Biotechnology – Principles and Processes Concepts Files |
| Biotechnology – Principles and Processes Master Files |
| Biotechnology – Principles and Processes Revision Note |
| Biotechnology – Principles and Processes Note 1 |
| Biotechnology – Principles and Processes Note 2 |
Biotechnology: Principles and Processes
Table of Contents
- Definition of Biotechnology
- Principles of Biotechnology
- Conceptual development of concepts of Genetic Engineering
- Tools of Recombinant DNA Technology
- Processes of Recombinant DNA Technology
- Frequently Asked Questions
Definition of Biotechnology
According to the European Federation of Technology, the term Biotechnology is defined as:
“The integration of natural science and organisms, cells, parts thereof, and molecular analogues for products and services.”
Principles of Biotechnology
Modern biotechnology is the result of the following two core techniques –
- Genetic Engineering – It includes several techniques that facilitate the alteration of genetic material, that is, RNA or DNA, in order to introduce them in host organisms. It includes changing of phenotype in host organism.
- In chemical engineering process, maintenance of microbial contamination free (sterile) atmosphere with an objective to initiate the large growth of desired eukaryotic or microbe cell in order to manufacture biotechnological products like enzymes, vaccines and antibiotics.
Conceptual development of Concepts of Genetic Engineering
Sexual reproduction is beneficial than asexual reproduction. This is so because sexual reproduction provides opportunity for formulation of new genetic set up and permits variation while asexual reproduction focuses on maintaining the genetic information. This idea plays the significant role in genetic engineering and has resulted in the creation of recombinant DNA, gene transfer and gene cloning. These processes have enabled the isolation of desirable gene without introducing undesirable one in the target organism.
When we focus on the construction of an artificial recombinant DNA molecule, it results the possibility of linking an encoding antibiotic resistance of a gene with an associated plasmid of Salomonella typhimurium. In 1972, Herbert Boyer and Stanley Cohen cut down the piece of DNA from plasmid and isolated the antibiotic resistance gene. This cutting of DNA was made possible by the discovery of molecular scissors (or restriction enzymes). The piece of DNA after being cut was linked or connected with plasmid DNA which acted as vectors to transfer the piece of DNA attached to it. An enzyme called DNA ligase (as it acts on cut DNA molecule and join there ends) made this linking possible and resulted in the development of in vitro and is referred as recombinant DNA.
Tools of Recombinant DNA Technology
The key tools of recombinant DNA technology are:
- Restriction Enzymes
- Polymerase Enzymes
- Ligases
- Vectors
- The Host Organisms
Restriction Enzymes
In order to limit the growth of bacteriophage in Escherichia coli, two enzymes were isolated in the year 1963, whereby first enzyme added methyl groups to DNA and was used to cut DNA (was termed as restriction endonuclease).
First Restriction Endonuclease – Hind II depended on the specific sequence of DNA nucleotide and always used to cut DNA molecule at a specific point after recognizing the base pairs sequence. This specific sequence of base pair is referred as recognition sequence. Besides Hind II, now we have around nine hundred restriction enzymes found from over 30 strains of bacteria. Each of these enzymes acknowledges different recognition sequences.
The restriction enzymes belong to larger class of enzymes called nucleases which are further classified as endonucleases and exonucleases. Endonucleases make cut at a specific position within DNA while exonucleases help in eliminating nucleotide from the end of DNA while,
Following diagram shows the steps in the formation of recombinant DNA. The restriction endonuclease enzyme – EcoRI plays an integral role in the formation, whereby each restriction endonuclease act by inspecting the length of DNA sequence. Once the specific recognition sequence is found, it binds with DNA and cut each of the strands of double helix at specific point in sugar phosphate backbone.

Palindromes – “Palindromes are the group of letters that form the same words when read both forward and backward, that is, MALAYALAM. Palindromes in DNA are the sequence of base pairs that reads same on the two strands when orientation of reading is kept same.”
Example of Palindromes:
The following sequence reads the same on two strands in 5’ → 3’ direction. This is again same if read in 3’ → 5’ direction.
5’ ——- GAATTC ——- 3’
3’ ——- CTTAAG ——- 5’
It is important to note that restriction enzymes cut DNA strand little far from the center of palindrome site. Added to this the cutting is carried out between the same 2 bases on opposite strands. This results in leaving of single stranded portion at the end. The overhanding stretches are called sticky end on each strand.
Restriction endonucleases are very useful in genetic engineering because it helps in forming ‘recombinant’ molecule of DNA (These molecules of DNA are composed of DNA from different genomes or sources).
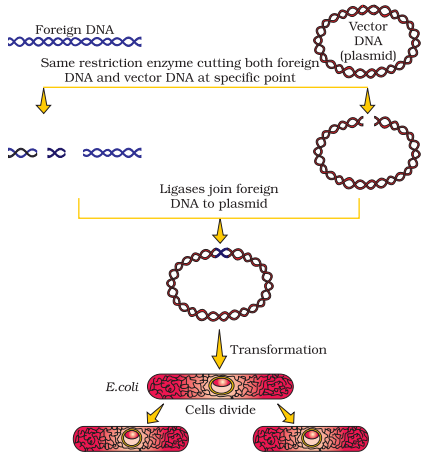
Following diagram represent the recombinant DNA technology. It is important to note that when removed or cut by the same restriction enzyme, the resultant DNA fragments possess same kind of “sticky ends” and these fragments are joined together via DNA ligases.
Separation and Isolation of DNA fragments
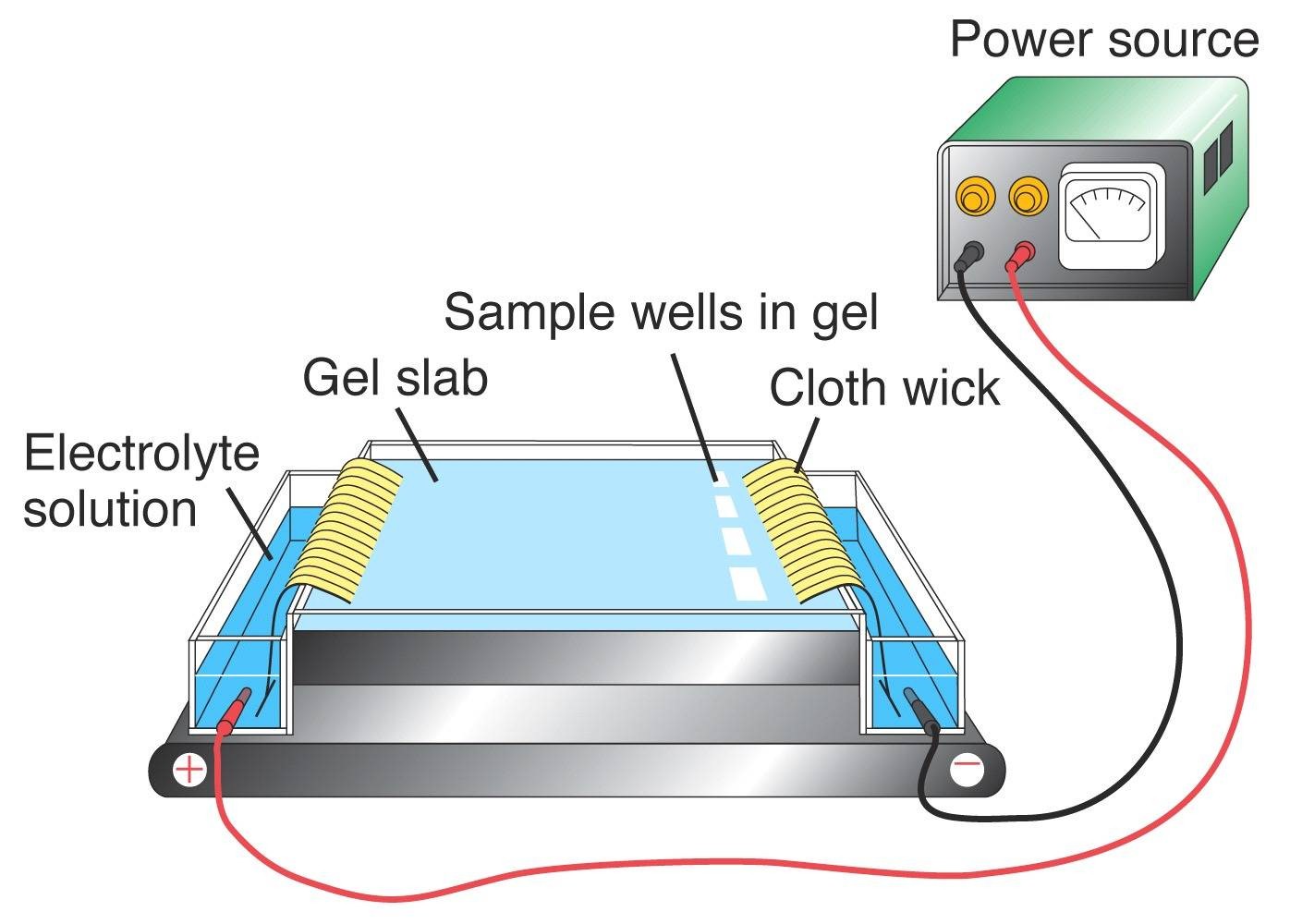
Restriction endonucleases form DNA fragments that are separated via technique gel electrophoresis. These DNA fragments are negatively charged molecule and is separated by moving them towards anode under an electric field via matrix or some medium. Most commonly used matrix is Agarose, a natural polymer extracted from sea weeds. The fragments of DNA are separated as per their size via sieving effect provided by agarose gel.
Following figure shows the gel electrophoresis instrumentation.
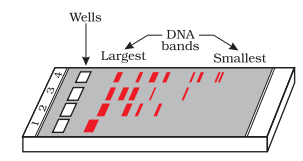
The separated DNA fragments are visible only after staining them with a compound called ethidium bromide.
Following figure shows the orange colored bands of DNA in ethidium bromide stained gel exposed to Ultra Violet light:
We all are aware of the fact that bacteriophages and plasmids have ability to replicate within bacterial cells and chromosomal DNA does not control this replication. Since, bacteriophages are very high per cell; they can easily copy their genome within bacterial cell. Added to this, plasmids have only 1 – 2 copies per cell. If we are able to connect an alien DNA piece with plasmid DNA or a bacteriophage, we are able to multiply its number equal to the copy number of plasmids and bacteriophage respectively. At present, vectors are engineered such that it facilitates easy linkage of foreign DNA along with selection of recombinants from non – recombinants.
Features required facilitating cloning into a vector
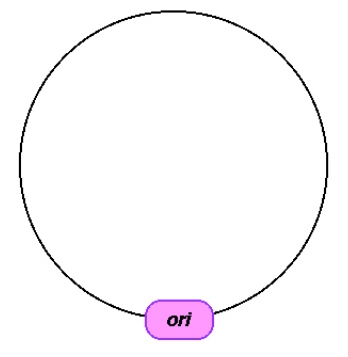
- Origin of replication – “It is the sequence where initiation or start of replication takes place and any DNA piece linked to this sequence is also replicated within host cells.” Origin of replication also helps in controlling the copy number of linked DNA, therefore, if anyone wants to recover several copies of target DNA, it must be cloned in vector whose origin support high copy number. Following image shows origin of replication of DNA.
- Selectable marker – Vector require selectable marker so as to eliminate and identify non – transformants and permit the growth of transformants on the selective basis. “Transformation is the process via which new DNA piece is introduced in host bacterium.” In the following figure, the green arrows signifies the selectable marker which permits the growth of transformants on selective basis.

- Cloning Sites – Recognition site are used to link alien DNA for the commonly used restriction enzyme. In case there is more than one recognition site in the vector, it results in the generation of several fragments that complicates the gene cloning.
- Vectors for cloning genes in animals and plants – Genes are transferred in plants and animals via viruses and bacteria. For instance, “Agrobacterioum tumifaciens, a pathogen of several dicot plants is able to deliver a piece of DNA called T-DNA to transform normal plant cells into tumor and direct these tumor cells to produce the chemical required by pathogen.” Similarly, in animals, retrovirus has the ability to transform normal cells in cancerous cells.
Competent Host (For Transformation with Recombinant DNA)
DNA is a hydrophilic molecule and therefore cannot pass through cell membrane. So, in order to force bacteria, to take up plasmid, the bacterial cells are first made “competent” via by treating them with specific concentration called divalent cation like calcium. Recombinant DNA is then forced into the cells by incubating cells on ice along with recombinant DNA.
This is not the only method of introducing alien DNA in host cells, rather a method called micro – injection as is directly injected into the nucleus of animal cells.
Processes of Recombinant DNA Technology
It includes several steps like:
- Isolation of DNA – DNA is enclosed in membrane; therefore, it is important to break the cells open in order to release DNA along with several other macromolecules like RNA, proteins, lipids and polysaccharides. This isolation process is carried out in the presence of several enzymes like cellulase (plant cells), lysozyme (bacteria) and chitinase (fungus).
- Cutting of DNA at specific locations – Restriction enzyme digestions are carried out by incubating purified DNA molecule with restriction enzyme in suitable conditions. Progression of restriction enzyme digestion is checked by Agarose gel electrophoresis. Since, DNA is charged negatively, it is attracted towards positive electrode and this entire process is repeated with the vector DNA as well.
Following is the diagrammatic representation of basic steps in recombinant DNA technology, using the bacterial plasmid as cloning vector. At initial stage, construction of recombinant DNA molecule takes place, then the cell is transported in host cells, followed by multiplication of recombinant DNA molecule, then division and finally the division results in a clone.

- Amplification of Gene of Interest using PCR – In Polymerase Chain Reaction (PCR), several copies of gene (or DNA) is formed in vitro taking help of two sets of primers. The enzyme with nucleotides extends the primers provided in reaction and the genomic DNA as template. Repetitive occurrence of this DNA replication, results in the amplification of segment of DNA to billion times. This repeated amplification is achieved by an enzyme thermostable DNA polymerase.
- Insertion of Recombinant DNA into the Host cells/ Organisms – Several methods are used to introduce litigated DNA into recipient cells after making them competent to receive and take up DNA present in its surroundings.
- Obtaining the Foreign Gene Product – While inserting the alien DNA piece in cloning vector, the multiplication of alien DNA takes place. In almost all recombinant technologies, the main objective is to produce desirable protein and therefore, it is important to express recombinant DNA. The foreign gene is expressed under appropriate conditions and the expression of this gene in host cells includes understanding of many technical details.
- Downstream Processing – After biosynthetic stage, the product is subjected through a series of processes like separation and purification, which aims at forming the finished product before marketing. These products are prepared with suitable preservatives and downstream process and quality control testing vary from product to product.
Frequently Asked Questions
Q1. Explain the contribution of Thermus aquaticus in the amplification of a gene of interest.
Sol. “Thermus aquaticus provides thermostable DNA polymerase, which can withstand high temperature required in denaturation and seperationf of DNA strands during PCR, i.e. Polymerase Chain Reaction.”
Q2. What are recombinant proteins? How do bioreactors help in their production?
Sol. “Recombinant proteins are produced by the expression of recombinant DNA in transgenic organisms.” Bioreactors help in developing recombinant proteins on large scale as large volumes of culture can be processed in bioreactors to produce desired quality of product and these have capability to provide optimum conditions of oxygen, pH, salts, substrates, etc.
Q3. What are bioreactors and how does it work?
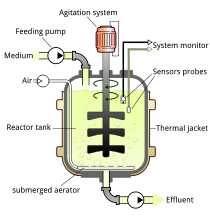
Sol. “Bioreactors are large vessels in which raw materials are biologically converted into specific products by microbes, plants and animals.” The reactors work by providing optimum condition to the process of culture by maintaining optimum temperature, oxygen, salt, pH and other required conditions. The two most commonly used bioreactors are Simple Stirred – tank bioreactors and Sparged Stirred – tank bioreactors.
Following figure shows the general structure of a continuous stirred tank type bioreactor:
Q4. Do eukaryotic cells have restriction endonucleases?
Sol. No, eukaryotic cells do not possess restriction endonucleases. This is so because all restriction endonucleases have been isolated from various strains of bacteria and are name as per genus and species. The first letter of enzyme comes from genus and second letter from species of prokaryotic cells from which they are isolated.
