Biodiversity can be assessed at three different levels. One of these is the genetic variation within each species.
(a) Outline two other levels at which biodiversity can be measured.
To calculate the genetic variation that exists within a species, scientists:
- obtain DNA sequences from many individuals of one species
- count the number of nucleotides that differ when the sequences of two individuals are compared
- repeat this with different pairs of individuals.
This allows scientists to calculate the mean number of differences at every nucleotide position along the sequence (mean number of nucleotide differences per site).
(b) Explain why scientists use databases and computers to calculate the mean number of nucleotide differences per site.
(c) Table 2.1 shows the mean number of nucleotide differences per site of some species.
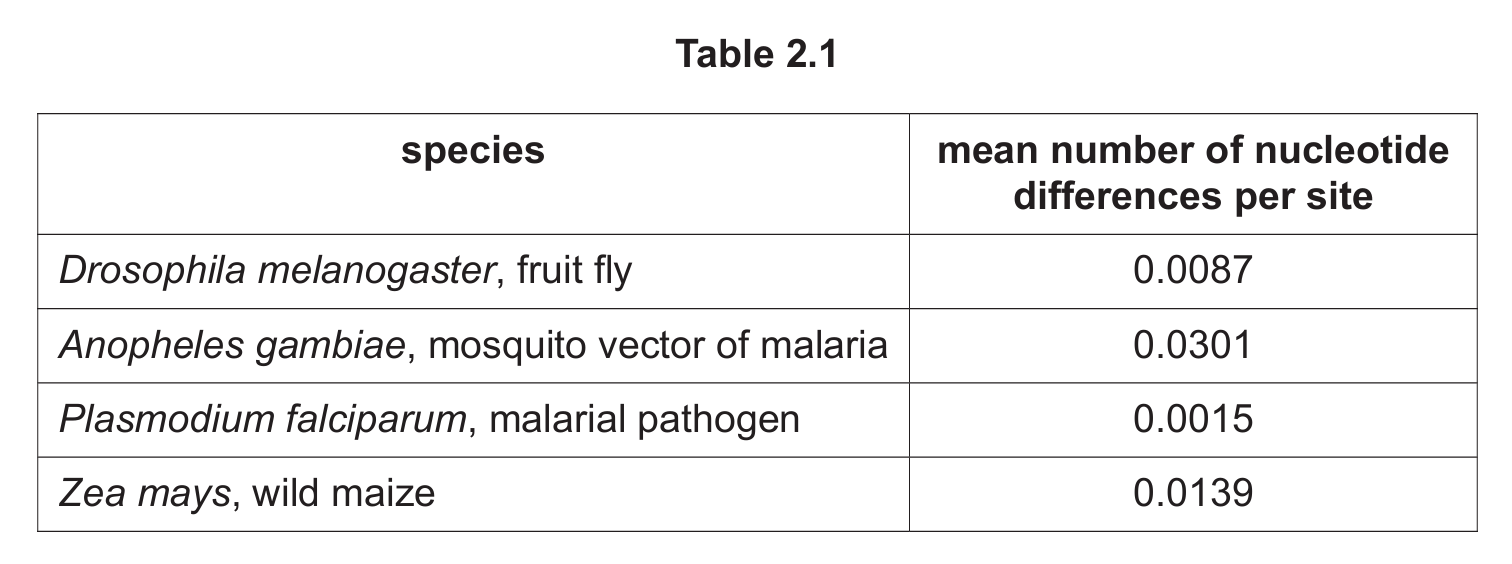
(i) State the genus name of the species that shows the most genetic variation.
(ii) State how many kingdoms of organisms are represented in Table 2.1.
(d) Genetic variation is considered important in the conservation of species. Low genetic variation is assumed to decrease the chance of the long-term survival of a species.
(i) Give reasons why low genetic variation may decrease the long-term survival of a species.
Fig. 2.1 shows how the International Union for the Conservation of Nature (IUCN) categorises species according to their conservation status.
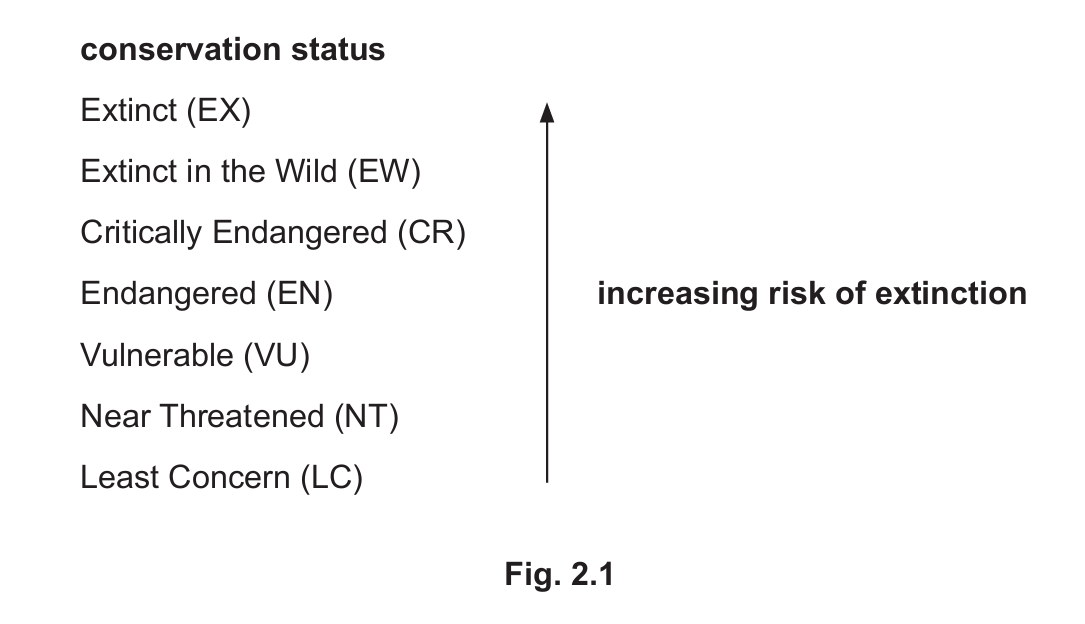
(ii) Question 2(d) states that ‘low genetic variation is assumed to decrease the chance of the long-term survival of a species’.
Predict the relationship between genetic variation and conservation status if this assumption is true.
(iii) Fig. 2.2 shows the mean number of nucleotide differences per site of some species and sub-species of mammal and their conservation status.
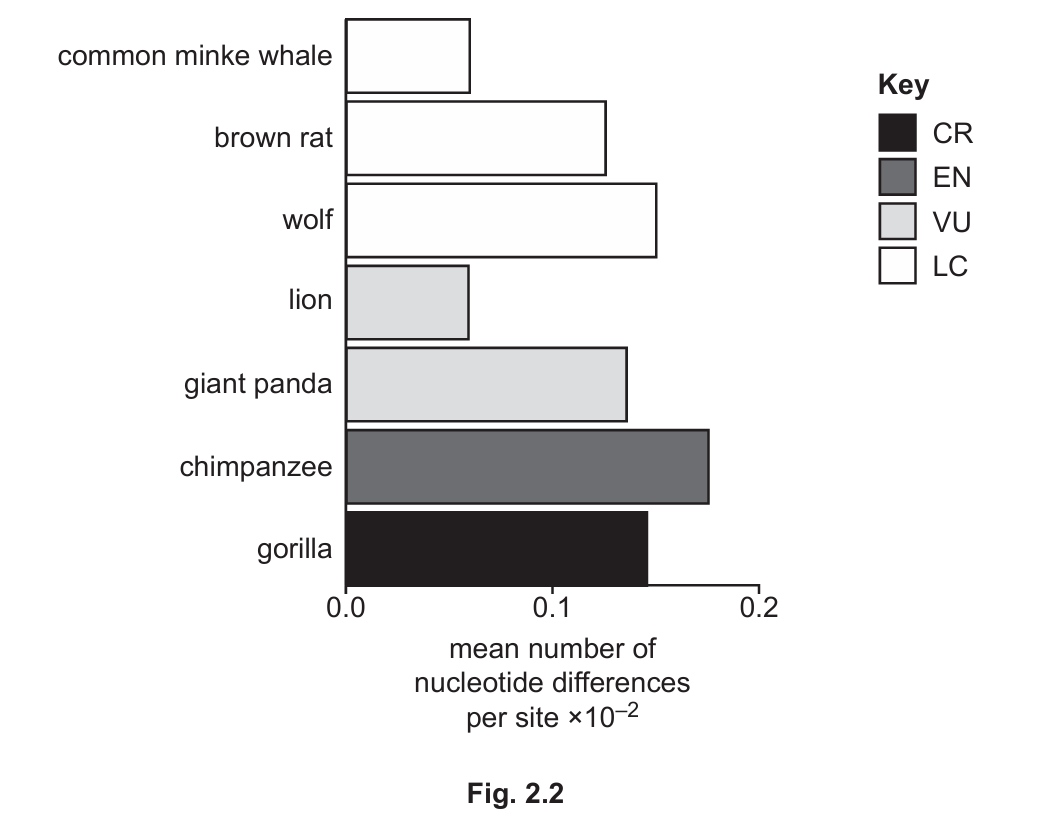
Assess whether the data in Fig. 2.2 provide support for the prediction you made in 2(d)(ii).
▶️ Answer/Explanation
(a)
1. Number of/different ecosystems/habitats in an area
2. Number of species and their relative abundance in an area
Explanation: Biodiversity can be measured at three levels: genetic diversity within species (already mentioned), species diversity (the variety and abundance of different species), and ecosystem diversity (the variety of different habitats and ecological processes).
(b)
1. To store/contain multiple/many genome/DNA/nucleotide sequences
2. To align/compare/search sequences efficiently
3. To process/analyze large quantities of data quickly
Explanation: Databases and computers are essential because DNA sequence analysis involves massive amounts of data that would be impractical to handle manually. Computers can quickly compare sequences from many individuals, identify differences, and calculate statistics like mean nucleotide differences per site.
(c)(i) Anopheles
Explanation: The mosquito Anopheles gambiae shows the highest mean number of nucleotide differences per site (0.0301), indicating the most genetic variation among the listed species.
(c)(ii) Three
Explanation: The table represents three kingdoms: Animalia (fruit fly, mosquito), Protista (malarial pathogen), and Plantae (wild maize).
(d)(i)
1. Less ability to adapt to changing environmental conditions
2. Fewer potentially beneficial alleles available for natural selection
3. Increased risk of extinction from diseases (all individuals equally susceptible)
Explanation: Genetic variation provides the raw material for evolution. Populations with low genetic variation have less capacity to adapt to environmental changes, making them more vulnerable to extinction. They may also suffer from inbreeding depression.
(d)(ii) Lower genetic variation means higher conservation status (greater risk of extinction)
Explanation: If the assumption is true, we would expect species with lower genetic variation to be classified in higher risk categories (like Endangered or Critically Endangered) on the IUCN Red List.
(d)(iii)
The data does not strongly support the prediction because:
1. Chimpanzees have high genetic variation but are Endangered
2. Minke whales have low genetic variation but are Least Concern
3. Gorillas have relatively high genetic variation but are Critically Endangered
Explanation: While genetic variation is important for conservation, the data shows that other factors (like habitat loss, hunting pressure) may be more significant in determining conservation status for these species. The relationship between genetic variation and extinction risk appears complex in this dataset.
