Question [Maximum marks: 11]
Penicillin is an antibiotic that interferes with the synthesis of cell walls in bacteria. Even before
penicillin became widely available in the 1940s, the enzyme penicillinase which breaks down
penicillin had been isolated. This enzyme is now found in many bacteria and gives them
resistance to penicillin.
Fig. 4.1 is a ribbon model of the structure of the enzyme penicillinase. The arrow indicates
the active site of the enzyme.
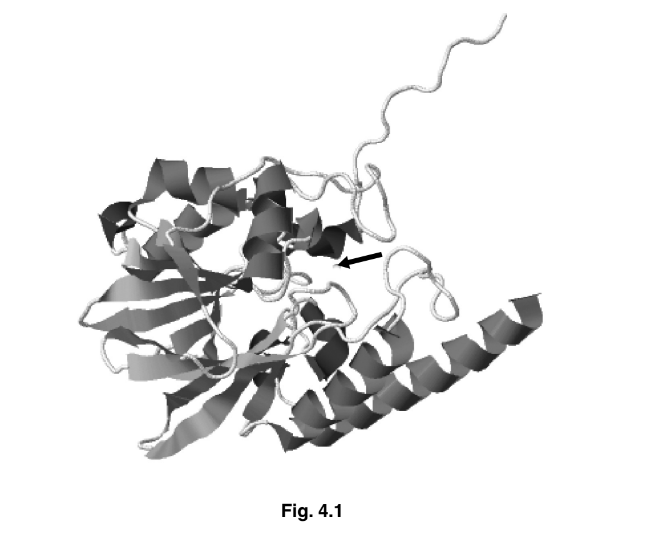
(a) Explain why the shape of the active site of an enzyme, such as penicillinase, is important.
(b) With reference to Fig. 4.1, identify the aspects of protein structure that are shown and
those that are not shown.
Fig. 4.2 shows the changes in energy during the progress of an uncatalysed reaction.
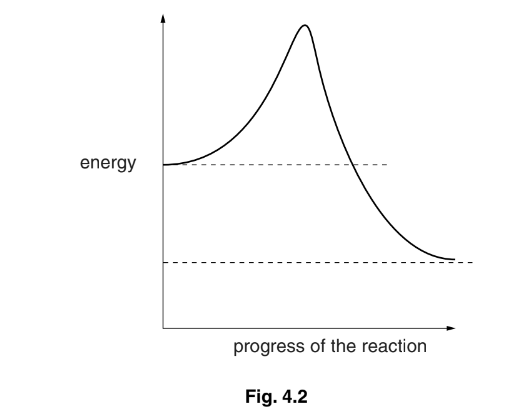
(c) (i) Draw on Fig. 4.2 a curve to show changes in energy during the progress of the
same reaction when catalysed by an enzyme.
(ii) State the term given to the energy level that must be overcome before a reaction can progress.
(d) Antibiotic resistance is a serious worldwide problem.
Suggest how antibiotics can be used effectively to avoid the development of widespread
resistance in bacteria.
Answer/Explanation
Answer: 4(a) this can be answered in the context of penicillinase
1 complementary shape ;
2 substrate, fits into / enters / binds to / with, active site ;
A enzyme-substrate complex / ESC
3 ref. to specificity ;
4 lock and key / induced fit ; A description of induced fit
5 ref. to temporary bonds form with, active site / R groups (of amino acid residues) ; [max 3]
4(b) shown to max 2
secondary structure ;
α / alpha, helix ; R ‘helix’ / helical structure unqualified by alpha
β pleated sheet ;
tertiary structure / folding ; ignore 3D shape or structure
globular ;
not shown to max 2
amino acids / primary structure / sequence of amino acids ;
(types of) R groups ;
bonds / named bonds ; A peptide
quaternary structure ;
prosthetic group ; [max 3]
4(c) (i) one lower peak inside line than uncatalysed ;
start and finish at, dotted lines / same energy levels as uncatalysed ;
(ii) activation (energy) / (energy of) activation ;
4(d) 1 do not prescribe for viral diseases ;
2 only use when necessary / do not overprescribe ;
3 only available on prescription / not available ‘over the counter’ ;
4 people must, complete the course / take as instructed ;
R take a long course
5 test to find out which is most appropriate antibiotic to use ;
A use most, appropriate / effective, antibiotics
A use narrow-spectrum antibiotics
6 details of sensitivity test ;
7 rotate / AW, antibiotics / use in combination ; R use many antibiotics
8 do not use same antibiotics for humans and animals ; [max 2]
Question
Fig. 3.1 shows the structure of the enzyme lysozyme.
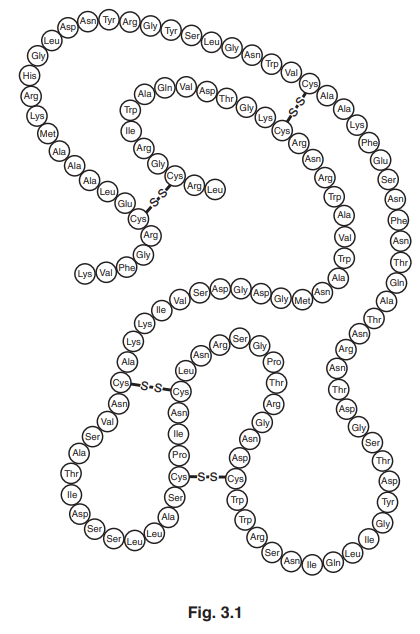
(a) (i) Name the two types of covalent bond in the structure in Fig. 3.1.[1]
(ii) The primary structure of lysozyme is shown in Fig. 3.1.
Explain the meaning of the term primary structure.[1]
(b) Lysozyme hydrolyses the β-1,4 glycosidic bonds present in compounds found in bacterial cell walls.
(i) State what is meant by the term hydrolysis.[1]
(ii) Suggest the type of biological molecule which is the substrate for lysozyme.[1]
(iii) Lysozyme uses the induced fit mechanism.
Explain the mode of action of an enzyme that uses the induced fit mechanism.[4]
(c) In human tears and saliva, lysozyme acts as an extracellular enzyme.
State what is meant by the term extracellular.[1]
(d) Fig. 3.2 shows the results of an investigation into the effect of substrate concentration on the rate of reaction catalysed by lysozyme.
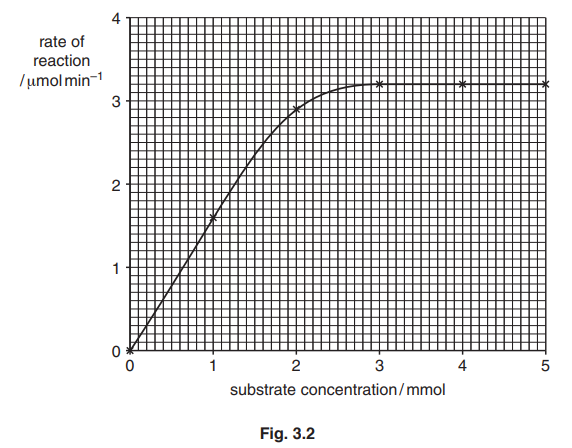
Use Fig. 3.2 to:
(i) state the lowest substrate concentration to give the maximum rate of reaction, Vmax.[1]
(ii) determine the Michaelis-Menten constant, Km.
Km = [1]
(e) The investigation was repeated in the presence of a competitive inhibitor of lysozyme.
Draw a curve on Fig. 3.2 to show the expected results. [2] [Total: 13]
Answer/Explanation
Ans:
3 (a) (i) peptide and disulfide ; R sulfide
(ii) sequence/ arrangement / order, of amino acids ; I ref. to disulfide bonds
(b) (i) breaking a (covalent) bond with addition of water ;
(ii) peptidoglycan / murein ; A carbohydrate / polysaccharide / amino sugar
(iii) four from
substrate shape not (exactly) complementary to active site shape /AW ;
active site (partially) flexible / changes shape slightly, when substrate,
enters /binds ;
(so) active site and substrate, now complementary / better fit ;
(allows) formation of enzyme-substrate complex ; A ES complex /ESC
AVP ; e.g. role of R-groups in active site interacting with substrate
lowers, activation energy /EA, so products form
(c) outside cells ; can be in a general context or in context of enzymes
(d) (i) 2.9 mmol ; A 2.75–3.0 mmol
(ii) 1 mmol ;
(e) single graph line with lower gradient ;
reaches or approaches plateau ;
Question
Catalase is an enzyme that catalyses the breakdown of hydrogen peroxide, a toxic waste product of metabolism.
Fig. 3.1 shows the results of an investigation into the effect of hydrogen peroxide concentration on the rate of the catalase-controlled reaction, with and without the presence of two different inhibitors.
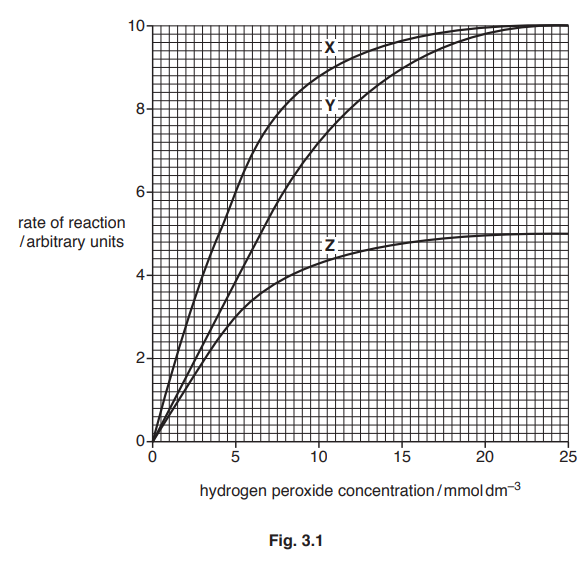
(a) The inhibitors used in the investigation have different modes of action.
Identify which of curves X, Y and Z are the results for:[1]
- the reaction with the non-competitive inhibitor
- the reaction with the competitive inhibitor
- the reaction without any inhibitor.
non-competitive inhibitor
competitive inhibitor
without any inhibitor
(b) With reference to Fig. 3.1, compare the maximum rate of reaction, Vmax, and the Michaelis-Menten constant, Km, for curves X, Y and Z.[4]
Vmax
Km
Hydrogen peroxide has a harmful effect on cells. One effect is to damage DNA.
(c) Describe the structure of DNA. [4]
(d) The cell has mechanisms to repair the damage to DNA caused by hydrogen peroxide. Errors in repair may cause a change to the structure of DNA.
Studies have investigated possible risks associated with foods and drinks that contain hydrogen peroxide. This is because the compound can be considered a mutagen. Mutagens cause mutations.
Explain why hydrogen peroxide can be considered a mutagen. [2] [Total: 11]
Answer/Explanation
Ans:
3(a) all three correct ;
with the non-competitive inhibitor Z
with the competitive inhibitor Y
without any inhibitor X
3(b) four from:
Vmax
1 X and Y same Vmax of 10 au ;
2 Vmax of, X/Y, higher than Z/ORA ; A (Vmax of), X/Y, 10 au v Z 5 au
A (Vmax of), X/Y, double the Vmax of Z
Km
3 X and Z same Km ; A Km of both is 4 mmol dm–3
4 X/Z, lower Km than Y/ORA ; A Km of, X/Z, 4mmol dm–3 v Y 6.5 mmol dm–3
5 reference to affinity for substrate ;
3(c) four from:
1 double helix ;
2 strands are held together by hydrogen bonds (between bases) ;
3 complementary base pairing / described as A-T and C-G ; A purine pairs with pyrimidine R thiamine
4 antiparallel stands / strands are 3’ to 5’ and 5’ to 3’ ; A strands run in opposite directions
5 (each strand has a sugar phosphate backbone with) phosphodiester bonds ;
6 (monomers / units /DNA) are (DNA) nucleotides /polynucleotide strands ;
7 (nucleotide = ) deoxyribose sugar, phosphate, nitrogenous (organic) base ;
A points from a diagram
3(d) two from:
1 idea that, hydrogen peroxide, damage / breaks, DNA and repair errors (may) occur ;
2 (so leads to) incorrect, nucleotide / base, inserted (during replication)/ change in, nucleotide / base, sequence (of DNA/RNA) ;
3 new allele (may be) formed ;
4 may result in an altered polypeptide /AW ;
