CIE AS/A Level Biology -6.2 Protein synthesis- Study Notes- New Syllabus
CIE AS/A Level Biology -6.2 Protein synthesis- Study Notes- New Syllabus
Ace A level Biology Exam with CIE AS/A Level Biology -6.2 Protein synthesis- Study Notes- New Syllabus
Key Concepts:
- state that a polypeptide is coded for by a gene and that a gene is a sequence of nucleotides that forms part of a DNA molecule
- describe the principle of the universal genetic code in which different triplets of DNA bases either code for specific amino acids or correspond to start and stop codons
- describe how the information in DNA is used during transcription and translation to construct polypeptides, including the roles of:
• RNA polymerase
• messenger RNA (mRNA)
• codons
• transfer RNA (tRNA)
• anticodons
• ribosomes - state that the strand of a DNA molecule that is used in transcription is called the transcribed or template strand and that the other strand is called the non-transcribed strand
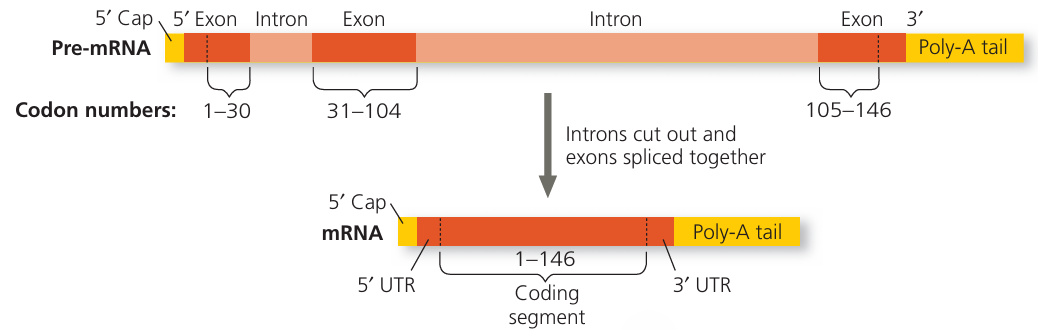
- explain that, in eukaryotes, the RNA molecule formed following transcription (primary transcript) is modified by the removal of non-coding sequences (introns) and the joining together of coding sequences (exons) to form mRNA
- state that a gene mutation is a change in the sequence of base pairs in a DNA molecule that may result in an altered polypeptide
- explain that a gene mutation is a result of substitution or deletion or insertion of nucleotides in DNA and outline how each of these types of mutation may affect the polypeptide produced
Genes and Polypeptides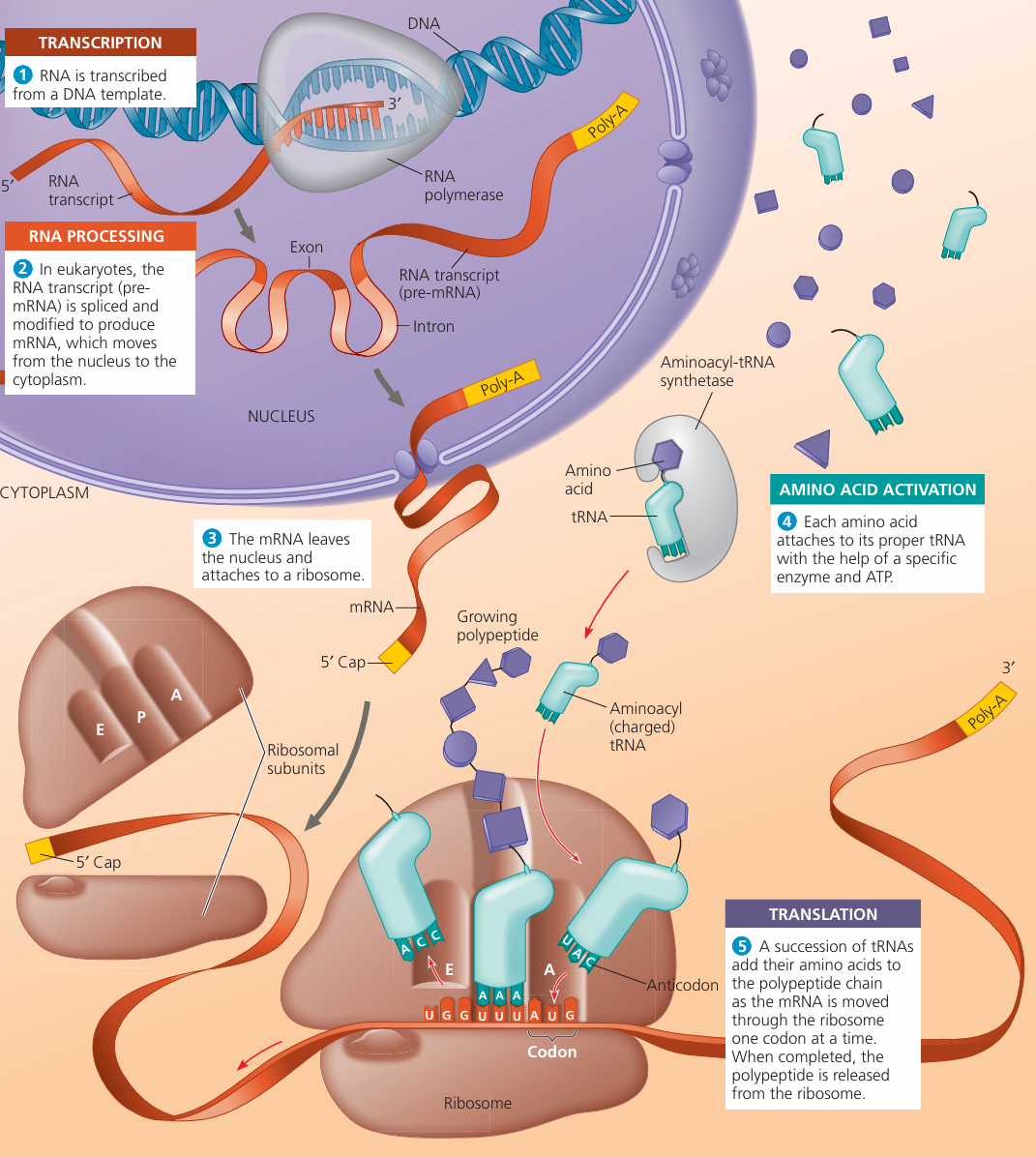
📌 Key Statement
- A polypeptide is coded for by a gene.
- A gene is a specific sequence of nucleotides that forms part of a DNA molecule.
- This nucleotide sequence contains the instructions for assembling a particular polypeptide (protein) by specifying the order of amino acids.
🔍 Explanation
- DNA stores genetic information in the form of triplet codes (three-nucleotide sequences called codons in mRNA after transcription).
- Each gene corresponds to a unique sequence of codons that determine the sequence of amino acids in a polypeptide.
- Central Dogma of Molecular Biology: DNA → mRNA → Polypeptide (via transcription and translation).
🧠 Example
- The insulin gene in humans contains a sequence of nucleotides that, when transcribed and translated, produces the insulin polypeptide (hormone regulating blood glucose).
Summary:
– A gene is a section of DNA made up of a specific nucleotide sequence.
– Each gene carries the instructions to make one polypeptide.
– Polypeptides are chains of amino acids whose sequence is determined by the order of nucleotides in the gene.
– A gene is a section of DNA made up of a specific nucleotide sequence.
– Each gene carries the instructions to make one polypeptide.
– Polypeptides are chains of amino acids whose sequence is determined by the order of nucleotides in the gene.
Universal Genetic Code
🌱 Definition
- The genetic code is the set of rules by which the sequence of bases in DNA (or mRNA) is translated into the sequence of amino acids in a polypeptide.
📌 Key Principles
- Triplet Code:
Three DNA bases (a triplet) correspond to one amino acid.
In mRNA, each triplet is called a codon.
Example: DNA triplet TAC → mRNA codon AUG → amino acid methionine. - Universal:
The genetic code is the same in almost all organisms (bacteria, plants, animals, humans).
A specific codon codes for the same amino acid regardless of the species.
Example: AUG codes for methionine in all living things. - Specific (Unambiguous):
Each codon specifies only one amino acid. - Degenerate (Redundant):
Most amino acids have more than one codon.
Example: GCU, GCC, GCA, GCG all code for alanine. - Start and Stop Codons:
Start codon: AUG → signals start of translation (codes for methionine).
Stop codons: UAA, UAG, UGA → signal the end of translation (no amino acid).
📊 Example Table of Codons
| mRNA Codon | Amino Acid | Function |
|---|---|---|
| AUG | Methionine | Start codon |
| UUU | Phenylalanine | Amino acid coding |
| UGC | Cysteine | Amino acid coding |
| UAA | – (no amino acid) | Stop codon |
| UAG | – (no amino acid) | Stop codon |
| UGA | – (no amino acid) | Stop codon |
Summary:
– Genetic code uses 3-base codons to specify amino acids.
– It is universal, specific, and degenerate.
– Start codon (AUG) begins translation; stop codons (UAA, UAG, UGA) end it.
– Universality is key evidence for the common origin of life.
– Genetic code uses 3-base codons to specify amino acids.
– It is universal, specific, and degenerate.
– Start codon (AUG) begins translation; stop codons (UAA, UAG, UGA) end it.
– Universality is key evidence for the common origin of life.
Protein Synthesis – Transcription & Translation
🌱 Overview
- Protein synthesis occurs in two main stages:
- Transcription – DNA → mRNA (nucleus)
- Translation – mRNA → polypeptide (cytoplasm, at ribosomes)
1️⃣ Transcription (in the Nucleus)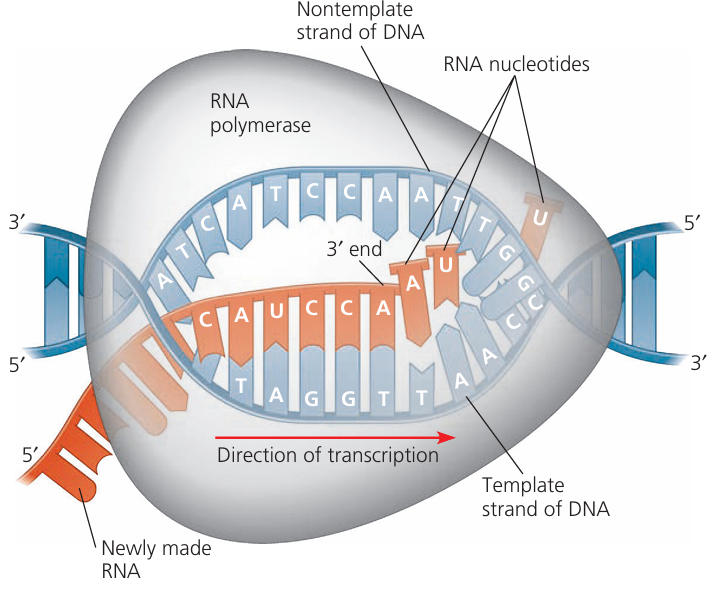
- Purpose: To produce a complementary mRNA copy of the gene.
- Steps:
- Initiation:
RNA polymerase binds to the promoter region of the gene.
DNA double helix unwinds, exposing the template strand. - Elongation:
RNA polymerase moves along the template DNA strand (3′ → 5′).
Matches RNA nucleotides to complementary DNA bases (A–U, T–A, G–C, C–G).
Forms messenger RNA (mRNA) with codons representing amino acids. - Termination:
RNA polymerase reaches a terminator sequence; mRNA detaches.
DNA helix reforms. - Post-transcriptional processing (in eukaryotes):
Introns removed, exons joined (splicing).
5′ cap and poly-A tail added for stability.
- Initiation:
2️⃣ Translation (at Ribosomes in Cytoplasm)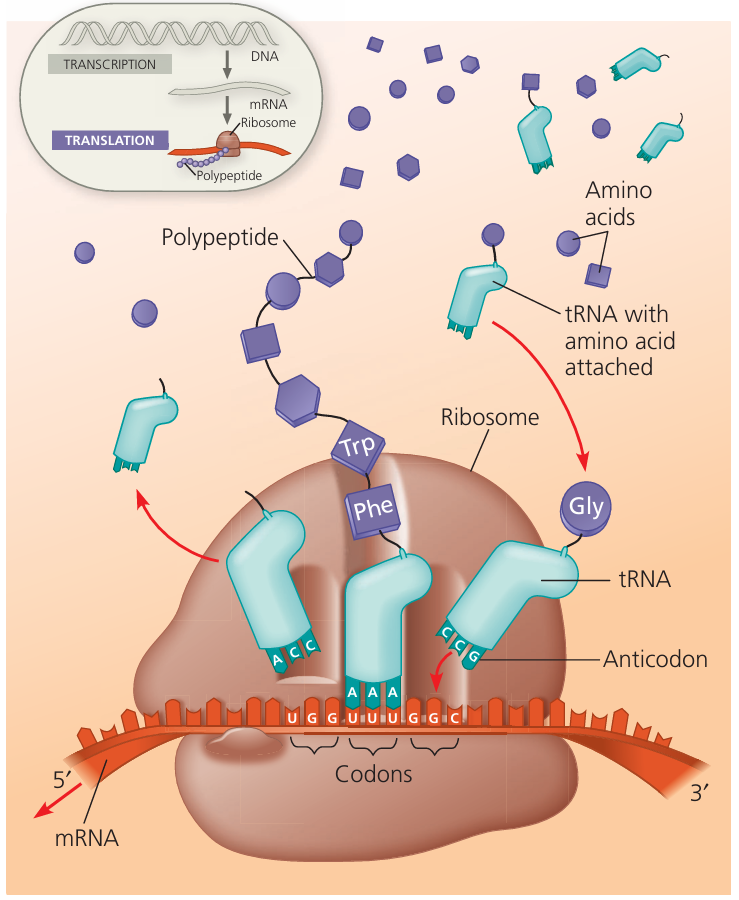
- Purpose: To use mRNA codons to assemble a polypeptide.
- Key Players:
- mRNA: carries genetic code from DNA to ribosome.
- tRNA (transfer RNA): delivers specific amino acids to ribosome; has an anticodon complementary to mRNA codon.
- Ribosome: site of protein synthesis; moves along mRNA, catalyses peptide bond formation.
- Steps:
- Initiation:
Ribosome binds to mRNA near the start codon (AUG).
First tRNA (anticodon UAC) carrying methionine binds. - Elongation:
Ribosome moves along mRNA, codon by codon.
Each codon is matched by a tRNA anticodon.
Amino acids linked by peptide bonds (via peptidyl transferase activity). - Termination:
Ribosome reaches a stop codon (UAA, UAG, UGA).
No tRNA matches → release factors detach ribosome, polypeptide released.
- Initiation:
📊 Roles of Key Components
| Component | Role |
|---|---|
| RNA polymerase | Catalyses mRNA synthesis from DNA template during transcription |
| mRNA | Carries genetic code (codons) from DNA to ribosome |
| Codons | Three-base sequences in mRNA that specify amino acids |
| tRNA | Delivers specific amino acids to ribosome |
| Anticodons | Three-base sequences on tRNA that pair with mRNA codons |
| Ribosomes | Assemble amino acids into polypeptides and catalyse peptide bond formation |
Summary:
– Transcription (nucleus) produces mRNA from DNA using RNA polymerase.
– mRNA codons are read by ribosomes during translation (cytoplasm).
– tRNA anticodons ensure correct amino acid sequence.
– Ribosomes link amino acids via peptide bonds to form a polypeptide.
– Transcription (nucleus) produces mRNA from DNA using RNA polymerase.
– mRNA codons are read by ribosomes during translation (cytoplasm).
– tRNA anticodons ensure correct amino acid sequence.
– Ribosomes link amino acids via peptide bonds to form a polypeptide.
DNA Strands in Transcription
🌱 Key Point
- The strand of DNA that is used as a template during transcription is called the transcribed strand or template strand.
- The other DNA strand (which has the same base sequence as the mRNA, except T instead of U) is called the non-transcribed strand or coding strand.
📌 Explanation
- Template (Transcribed) Strand:
- Runs in the 3′ → 5′ direction so RNA polymerase can synthesise mRNA in the 5′ → 3′ direction.
- mRNA bases are complementary to this strand.
- Non-Template (Non-Transcribed / Coding) Strand:
- Runs in the 5′ → 3′ direction.
- Has the same sequence as mRNA (except DNA has T, RNA has U).
- Not used for direct mRNA synthesis.
Summary:
– Template strand = transcribed strand (complementary to mRNA).
– Non-transcribed strand = coding strand (same sequence as mRNA except T instead of U).
– Template strand = transcribed strand (complementary to mRNA).
– Non-transcribed strand = coding strand (same sequence as mRNA except T instead of U).
RNA Processing in Eukaryotes
🌱 Key Point
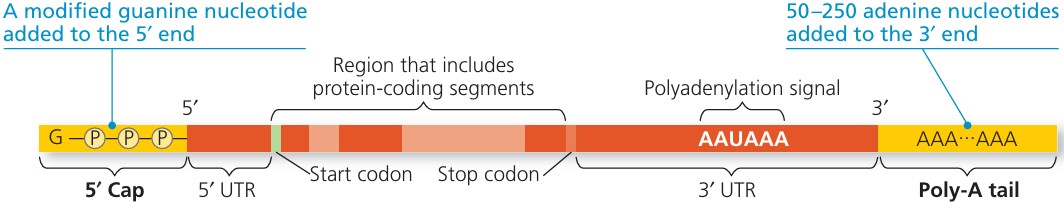
- In eukaryotic cells, the RNA formed immediately after transcription is called the primary transcript (or pre-mRNA).
- This primary transcript cannot be used directly for protein synthesis—it must be processed into mature mRNA.
🔬 Steps in RNA Processing
- Splicing – removal of introns
- Introns = non-coding sequences of DNA/RNA that do not code for amino acids.
 Removed by a complex called the spliceosome.
Removed by a complex called the spliceosome.
- Joining of Exons
- Exons = coding sequences that contain instructions for making a protein.
- After introns are removed, exons are joined in the correct order to produce a continuous coding sequence.
- Additional Modifications (for stability and translation)
- 5′ cap: Modified guanine nucleotide added to the 5′ end—protects mRNA from degradation and helps ribosome binding.
- Poly-A tail: Chain of adenine nucleotides added to the 3′ end—increases stability.
📌 Example
- Gene sequence: Exon 1 – Intron – Exon 2 – Intron – Exon 3
- After RNA processing → Exon 1 – Exon 2 – Exon 3 (mature mRNA)
| Term | Meaning |
|---|---|
| Primary transcript (pre-mRNA) | RNA directly after transcription, contains both exons and introns |
| Introns | Non-coding sequences removed during RNA processing |
| Exons | Coding sequences joined to form mature mRNA |
| Mature mRNA | Processed RNA with only exons, ready for translation |
Summary:
– Transcription produces pre-mRNA with both exons and introns.
– Introns are removed, exons are joined (splicing).
– Additional modifications (5′ cap and poly-A tail) produce stable, mature mRNA for translation.
– Transcription produces pre-mRNA with both exons and introns.
– Introns are removed, exons are joined (splicing).
– Additional modifications (5′ cap and poly-A tail) produce stable, mature mRNA for translation.
Gene Mutation
🌱 Definition
- A gene mutation is a change in the sequence of base pairs (nucleotides) in a DNA molecule.
- This change can alter the genetic code and may affect the sequence of amino acids in the resulting polypeptide.
🔬 Possible Effects
- Altered polypeptide → mutation changes the codon(s), leading to a different amino acid being incorporated into the protein.
- No change → some mutations are silent (due to the degeneracy of the genetic code).
- Loss of function → protein becomes non-functional due to major changes in structure.
- Gain of function → rare cases where a mutation gives a protein a new or enhanced function.
| Term | Meaning |
|---|---|
| Gene mutation | Permanent change in the DNA base sequence of a gene |
| Point mutation | Single base change (substitution, insertion, deletion) |
| Silent mutation | Change in base sequence with no effect on amino acid sequence |
| Missense mutation | Change in one amino acid in the polypeptide |
| Nonsense mutation | Change results in a premature stop codon |
Summary:
– Gene mutation = change in DNA base sequence.
– May cause changes in the polypeptide produced.
– Effects vary: no change, altered function, or complete loss of function.
– Gene mutation = change in DNA base sequence.
– May cause changes in the polypeptide produced.
– Effects vary: no change, altered function, or complete loss of function.
Gene Mutations – Types & Effects
🌱 Definition Recap
- A gene mutation is a change in the sequence of nucleotides in a DNA molecule.
- It can occur due to substitution, deletion, or insertion of nucleotides.
- Such changes may alter the mRNA codons and, in turn, the polypeptide sequence.
🔬 Types of Gene Mutations & Their Effects
| Type of Mutation | Description | Possible Effect on Polypeptide |
|---|---|---|
| Substitution | One nucleotide is replaced by another | – May change a codon to one coding for a different amino acid (missense mutation). – May change a codon to a stop codon (nonsense mutation), producing a shorter, non-functional polypeptide. – May have no effect (silent mutation) if the codon still codes for the same amino acid. |
| Deletion | One or more nucleotides are removed from the sequence | – If not in multiples of 3 bases, causes a frameshift mutation → shifts the reading frame → changes every amino acid after the deletion. – Often results in a completely non-functional protein. |
| Insertion | One or more extra nucleotides are added to the sequence | – If not in multiples of 3 bases, also causes a frameshift mutation → major change in amino acid sequence. – Usually produces a non-functional or truncated protein. |
📌 Key Points
- Frameshift mutations (caused by most insertions/deletions) are usually more harmful than single base substitutions.
- Substitution mutations can vary from harmless to severely damaging depending on where they occur.
- Mutations can be spontaneous (errors during DNA replication) or induced (by radiation, chemicals, viruses).
Summary:
– Gene mutations occur by substitution, deletion, or insertion of nucleotides.
– Substitution: may cause missense, nonsense, or silent mutations.
– Insertion/Deletion: often cause frameshift → drastic protein changes.
– Gene mutations occur by substitution, deletion, or insertion of nucleotides.
– Substitution: may cause missense, nonsense, or silent mutations.
– Insertion/Deletion: often cause frameshift → drastic protein changes.