Question
The following figure is a cladogram that represents the suspected ancestry of five species.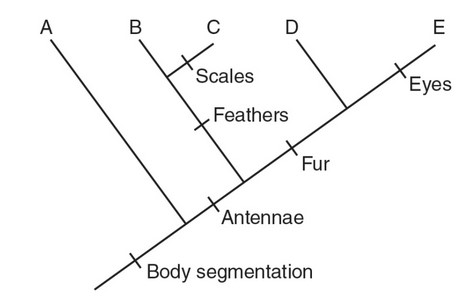
(a) Describe the characteristic(s) of the species that is the outgroup on this cladogram.
(b) Describe the similarities and differences in the characteristics present in species B and species D.
(c) A new species (X) is discovered, which has body segmentation, antennae, and fur but does not have eyes or scales. Construct a line that represents the ancestry of species X on the cladogram.
(d) Explain why species C is placed off of the same branch as species B and not the same branch as species D.
▶️Answer/Explanation
Ans:
(a) Species A is the outgroup, and according to the cladogram, the only
characteristic it possesses is body segmentation.
(b) Species B has body segmentation, antennae, and feathers. Species
D has body segmentation, antennae, and fur.
(c)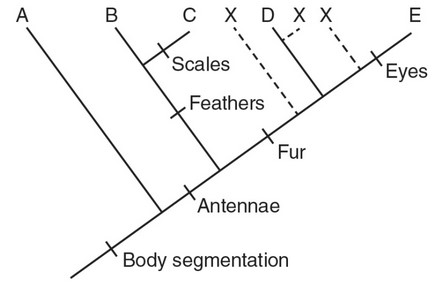
(All three possible positions for species X on the above cladogram
would be considered acceptable.)
(d) Species C has feathers, which is a characteristic that it has in
common with species B. Species D has fur, which species C does
not possess. Therefore, species C is placed off of the same branch
as species B and not on the same branch as species D.
Question
Refer to the following table and phylogenies.
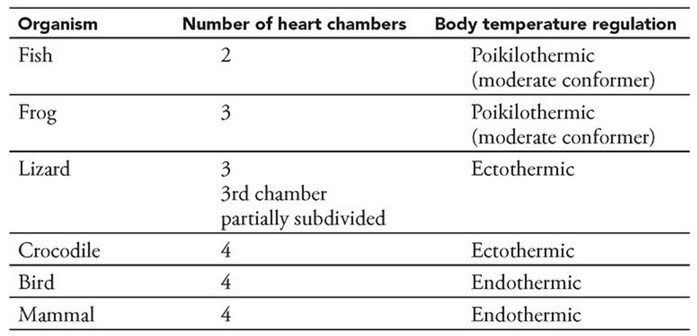
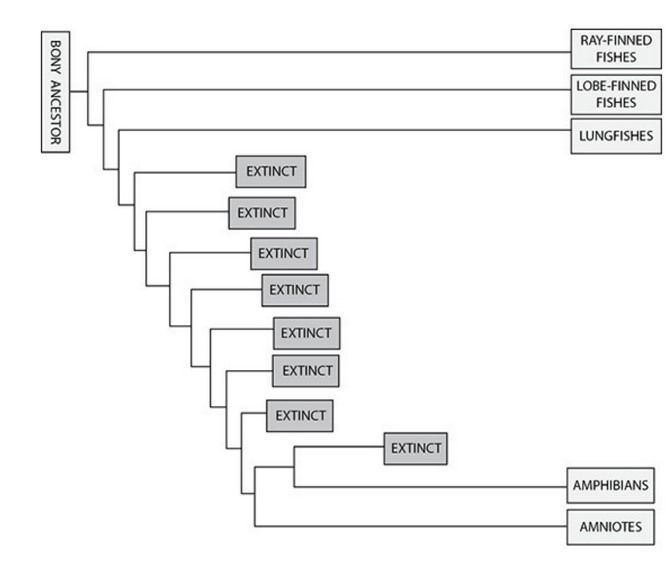
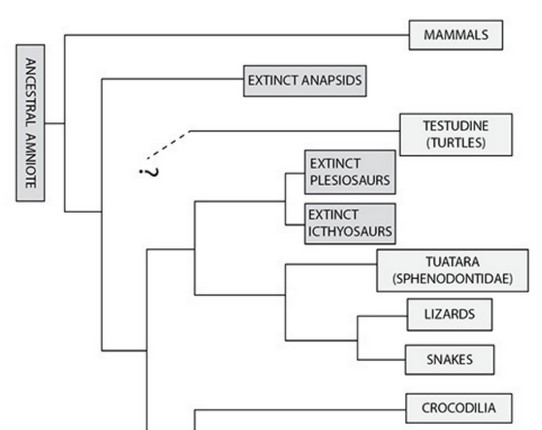
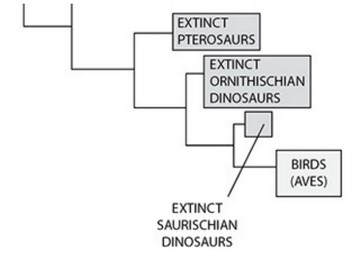
Construct a hypothesis about the evolution of endothermy. The hypothesis should address whether endothermy is homologous (i.e., it evolved once) or analogous (i.e., it evolved independently at least twice) in birds and mammals. Refer to the table and phylogenies to support your claim.
▶️Answer/Explanation
Ans:
Mammals and birds arose from different reptilian lineages, so endothermy likely evolved separately (independently) in the two classes, and its evolution in birds and mammals is probably analogous. The common ancestor between the birds and the mammals was too long ago for endothermy to be an ancestral trait in birds. However, the mode of body temperature regulation in Saurischia is not known.
Question
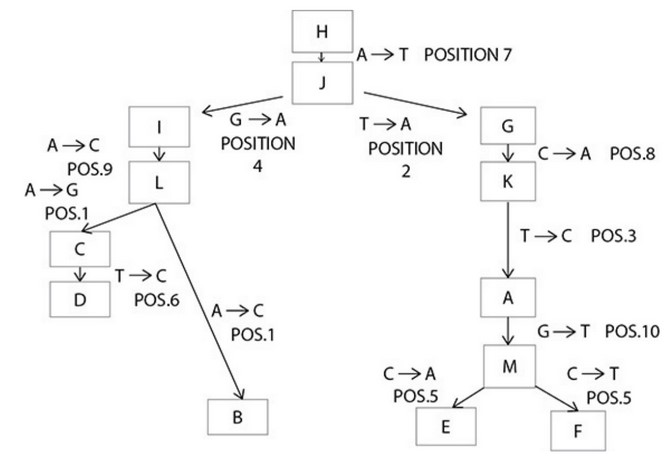
Refer to the following sequence data.
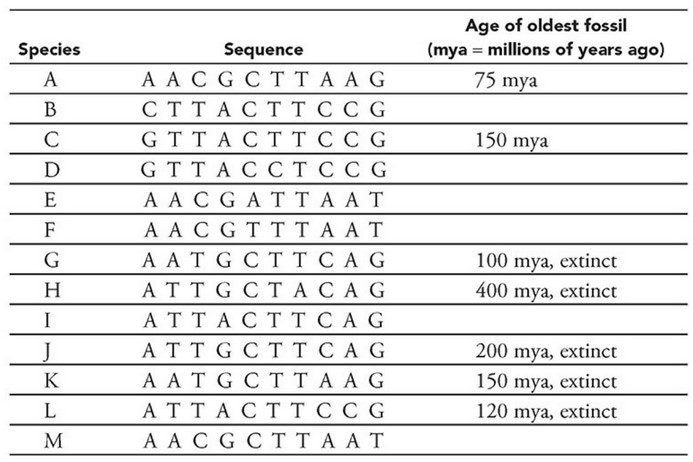
- Construct a phylogeny using the preceding sequence data.
- State three assumptions you used in constructing the phylogeny.
▶️Answer/Explanation
Ans:
Phylogenies are hypotheses that attempt to explain the evolutionary
relationship among organisms. Several types of data are used to construct a
phylogeny, but sometimes it is useful to construct two or more phylogenies,
each of which is based on only one type of data, in order to compare the
different predictions made by the different types of data.
No one knows exactly what happened from 3.8 billion years ago to now,
so everyone’s reconstruction of life’s history is a hypothesis. A good
hypothesis is consistent with the data available and provides testable
predictions.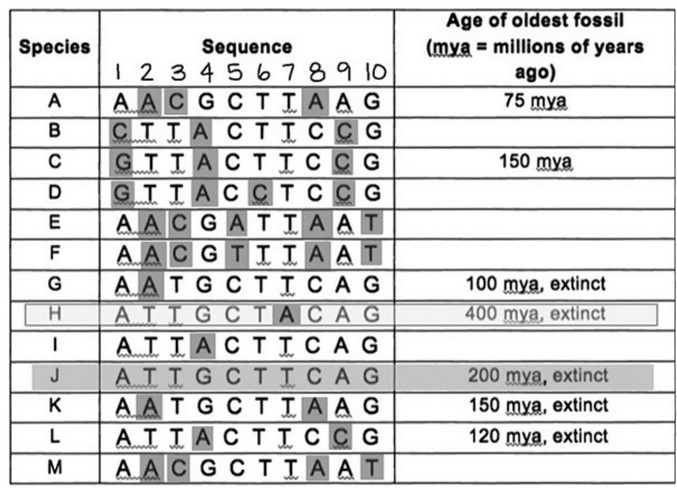
To handle sequence data (refer to the preceding sequence data and the
following phylogeny):
(1) Evaluate sequences by position number. For example, vertically
scanning position 1 reveals that A is more common than G or C.
(2) Mark nucleotide differences so they can be tallied later. Sequences
G, H, I, and J differ by only one nucleotide (they each have only one
mark), so one (or more) of them is the ancestral species.
Assumption 1: Species H is ancestral. The oldest known fossil for
species H is 400 million years old (myo). There’s no fossil data for species
I. Fossil data for species J and G are <400 myo. The evidence supporting H
as the ancestral sequence is weak.
The process involves some trial and error, so expect to reconfigure your
phylogeny a few times.
(1) Find the sequence with the least number of nucleotide
differences with the ancestral species. H and J have one difference
at position 7. All other species have a T in that position, so a
reasonable hypothesis is that H is ancestral to J, and J gave rise to
the other species. Remember that this is just a story until there is
sufficient data to support the hypotheses.
(2) Link sequences in “time” by locating sequences that differ in
only one (maybe two) positions, particularly if it is a change in
the same position. The sequences that are most similar to J are I
and G, which each differ from J by only one nucleotide, but the
differences are present at different positions (position 4 in I and
position 2 in G). This could have resulted from a few different
histories, one solely due to divergence, and another that includes
divergence and a back mutation.
Assumption 2: no back mutations. A back mutation is when a mutation
occurs and then in later generations, another mutation reverses the original.
Many sequence phylogenies assume no back mutations (at first).
(1) Branch points may be identified by single substitutions in
descendant species that occur at different sequence positions
from the common ancestor. The sequences between ancestor
species J and descendants I and G differ by one nucleotide but at
two different positions (position 4 in I and 2 in G).
(2) Branch points may be identified by a different nucleotide
substitution in the same position. The sequence from species L
differs from species C and B at the same position but at a different
location (position 1 changed to a G in species C and a C in species
B). The sequence from species M differs from species E and F at position 5. In species E, C was substituted with A, and in species F,
C was substituted with T.
Assumption 3: A different, single nucleotide change at the same
position indicates the divergence of two populations from the common
ancestor. It is possible that the sequence change occurred in the transition
from M to E (or F) and then another substitution occurred at the same
location from the divergence of F from E (or E from F).
The following phylogeny shows the nucleotide and position numbers for
each change. It is not the only phylogeny that makes sense from the
data. There are also numerous assumptions made in the construction of
the phylogeny that were not specifically listed, such as the assumption
that the sequences with the least number of differences in this specific piece
of sequence are more closely related and that nucleotide substitutions
always indicated divergence.