A.Bioenergetics
➢ Glucose, starch, and fat all energy-rich, but the bonds must be broken in order for the energy to be released
➢ First Law of Thermodynamics: Energy cannot be created or destroyed. The sum of energy in the universe is constant.
➢ Second Law of Thermodynamics: Energy transfer leads to less and less organization. The universe tends towards entropy
➢ Types of Reactions
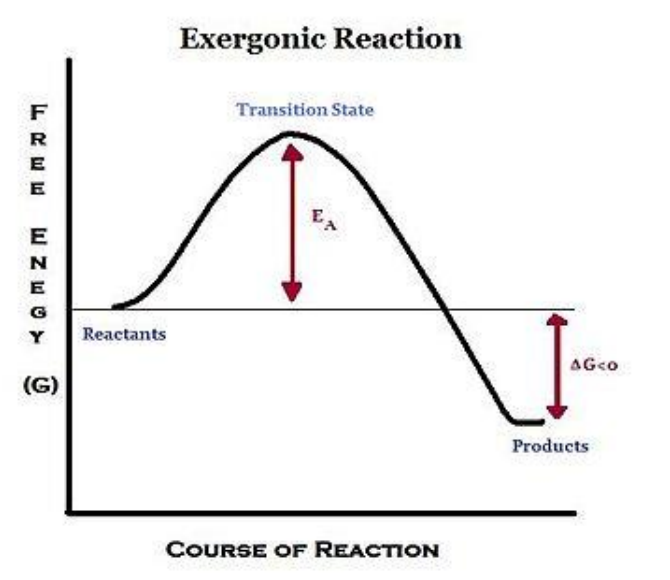
- Exergonic
■ Products have less energy than the reactants
■ Energy is given off during the reaction
■ Ex. oxidation of molecules in mitochondria
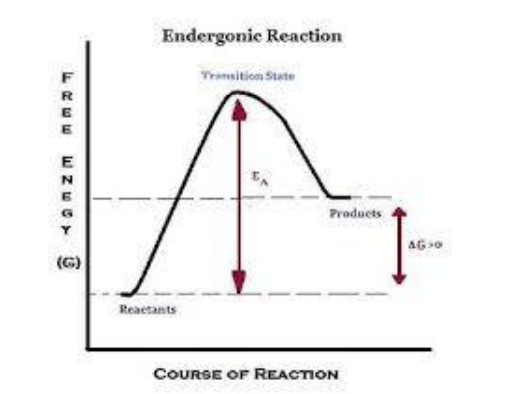
- Endergonic
■ Require an input of energy
■ Products have more energy than reactants
■ Ex. plants’ use of $CO_2$ and water to form sugars
B. Gibbs Free Energy
➢ $ΔG=ΔH-TΔS$
- T=temperature
- H=enthalpy (measure of energy in a thermodynamic system)
- S=entropy
- Change in the Gibbs free energy of a reaction determines whether the reaction in
favorable (spontaneous, negative) or unfavorable (nonspontaneous, positive) - Used to figure out if, without adding energy, the reactants will stay as they re or be converted to products
➢ Spontaneous Reactions
- Occur without a net addition of energy
- $ΔG<0$=exergonic
- $ΔG>0$=endergonic
■ Only occur if energy is added
➢ Activation Energy - Even though exergonic reactions release energy, the reaction still needs energy to start off with
■ Reactants must first go into transition state before turning into products
■ Activation energy is the energy needed to achieve the transition state
■ Bonds must be broken before new bonds can form
C. Enzymes
➢ Biological catalysts that speed up reactions
- Accomplished by lowering activation energy and helping transition state form
➢ Lowers activation energy by:
- Orienting substrate correctly
- Straining substrate bonds
- Providing favorable microenvironment
- Bonding to substrate
➢ Do NOT change the energy of the starting point or the ending point of the reaction. Only lower activation energy
➢ Enzyme Specificity
- Each enzyme catalyzes only one kind of reactions
- Enzyme are usually named after the molecules they target
■ Replace suffix of substrate with -ase
● Ex. maltose catalyzed by maltase - Substrates are the targeted molecules (reactant)
➢ Enzyme-Substrate Complex
- Enzyme brings about transition state by helping the substrate(s) get into position
- Accomplished through active site
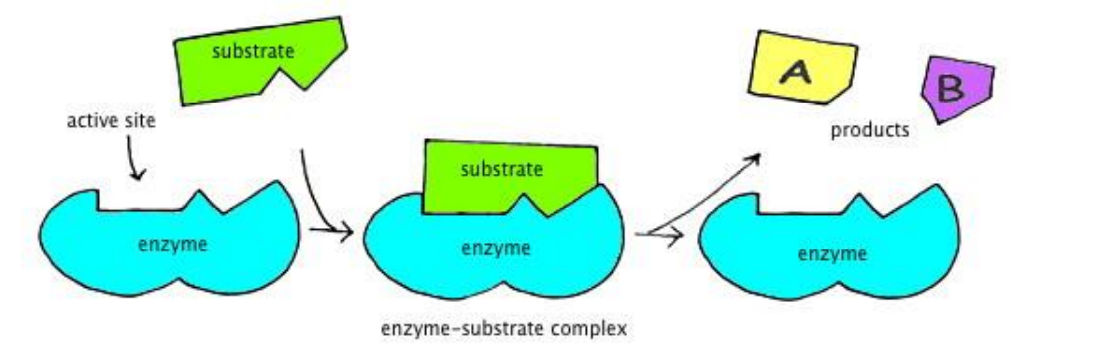
- Once the reaction has occurred, the enzyme is released from the complex and restored to its original state
■ Now the enzyme is free react with other substrates again - Induced fit
■ Enzyme slightly changes shape to accommodate the shape of substrates
■ Sometimes certain factors are necessary for this process
■ Cofactors sometimes aid induced fit and also help catalyze reactions
● Nonprotein helpers of enzymes
● Ex. vitamins
➢ Factors affecting reaction rates
○ Temperature
■ Although the rate of reaction increases with temperature, it only does so up to a point, because too much heat can denature an enzyme
■ $Q_10$
● Measure of sensitivity of a physiological process of enzymatic reaction rate
$Q_{10}=\left(\frac{R_2}{R_1}\right)^{\left(\frac{10}{T_2-T_1}\right)}$
- Temp must be celsius or kelvin
■ Same unit for $T_1 and T_2$ - Two reaction rates $(k_1 and k_2) $must have same unit
- Reaction rates with$ Q_10=1$ are temperature independent
- pH
■ Most enzyme’s ideal $\text pH$ is 7

➢ Enzyme Regulation
- Cell can control enzymatic reactions by regulating the conditions that change the shape of the enzyme
- Can be turned off/on by things that bind to them
■ Some bind at active site
■ Some bin at allosteric sites (non-active sites) - Competitive inhibition
■ If a substance has the exact complementary shape to the active site, it can compete with the substrate and block it from getting into the active site
■ If there is enough substrate available, it will out-compete the inhibitor and the reaction will occur
■ As substrate is used up, inhibitor out-competes the substrate and less reaction will occur - Allosteric inhibitors/Non Competitive inhibition
■ Binds to an allosteric site
■ Distorts shape of enzyme so it cannot function until the inhibitor is removed
■ Substrate can still bind if active site is intact, but the enzyme will not be able to catalyze the reaction
■ Activators can also be used to stabilize the enzyme’s active state
■ Inhibitors stabilize the inactive state

- Enzymes can also be controlled by negative feedback mechanisms
■ Product of reaction the enzyme is helping is also an allosteric inhibitor
■ Prevents a cell from wasting resources by synthesizing more of a product than is needed
D. Reaction Coupling
➢ ATP consists of a molecule of adenosine bonded to 3 phosphates
- Carries enormous amount of energy within phosphate bonds
➢ When a cell needs energy, it splits off the 3rd phosphate, forming adenosine diphosphate (ADP) and one loose phosphate $(P_i)$ in the process
- $ \text{ATP→ADP + Pi + energy}$
➢ ATP is relatively neat and easy to form
➢ Sources
- Cellular respiration
■ Sugar turned into ATP
● In plants, sugar is made by photosynthesis
● In animals, sugar is taken from food consumed
E. Photosynthesis
➢ $6CO_2 + H_2O → C_6H_12O_6 + 6O_2$
➢ Chloroplast structure
- Stroma=inner fluid-filled region
- grana=structures inside stroma that look like stacks of coins
- thylakoids=”coins” of grana
■ Contain chlorophyll, a light-absorbing pigment that drives photosynthesis
● Chlorophyll a
● Chlorophyll b
● Carotenoids
● Pigments gather light, but are not able to excite electrons, only one molecule in the reaction center can
■ Contains enzymes involved in photosynthesis
➢ 2 reaction centers:
- Photosystem I (PS I)
■ $p700$ - Photosystem II (PS II)
■ $P680$ - Both comprised of a Light harvesting complex, where a photon of light is passed like a wave between pigments and a Reaction center complex, which contains chlorophyll-a and uses light energy to “boost” and electrons and pass onto primary electron acceptor
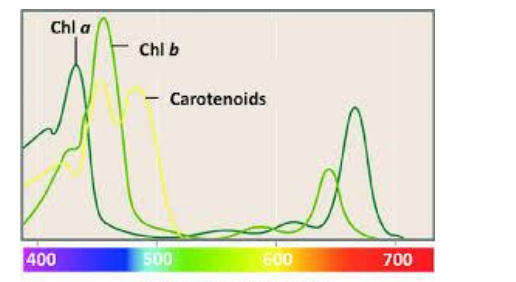
➢ Absorption spectrum measures how well a certain pigment absorbs electromagnetic radiation
- Opposite of emission spectrum
- Chlorophyll a and b absorb blue and red light but reflect green (reason why plants are usually green)
- Carotenoids absorb light at blue-green end, and reflect red light
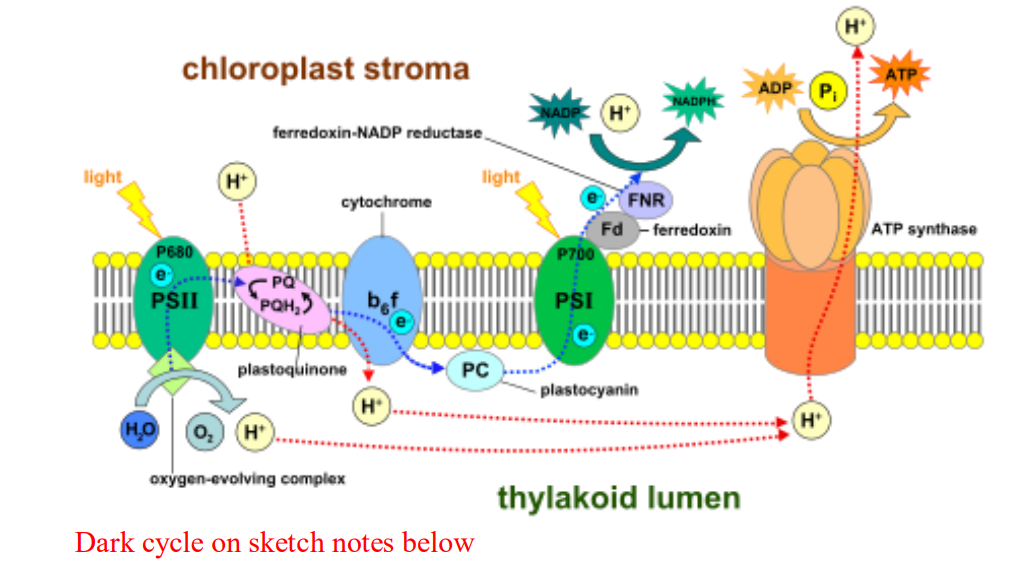
➢ Light reactions
- When a leaf captures sunlight, the energy is sent to$ p680$ of photosystem II
■ Sidenote: it may seem weird that the light reaction starts off in PSII and not PS I but its only called PS I because it was discovered first - Activated electrons trapped by p680 and passed down to molecule called the primary acceptor, and then they are passed down to carriers in the electron transport chain
- Photolysis
■ To replenish electrons in the thylakoid, water is split into $O^{-}$ , $2H^{+}$ , and electrons
● Water is split again into hydrogen ions (used for ETC) and Oxygen (released) - As the energized electrons from PSII travel down the ETC, they pump H+ into the thylakoid lumen
■ Proton gradient is created
● As hydrogen ions move back into the stroma through ATP synthase along their concentration gradient, ATP is created - After the electrons leave PS II, they enter PSI, where they are passed through a second ETC until they reach the final electron acceptor$ NADP^{+}$ to make $NADPH$
F. Cellular Respiration
➢$ C_6H_12O_6 + 6O_2 ⟶6CO_2 + 6H_2O + ATP$
➢ Aerobic respiration: ATP made in the presence of oxygen
➢ Anaerobic respiration: ATP made without presence of oxygen
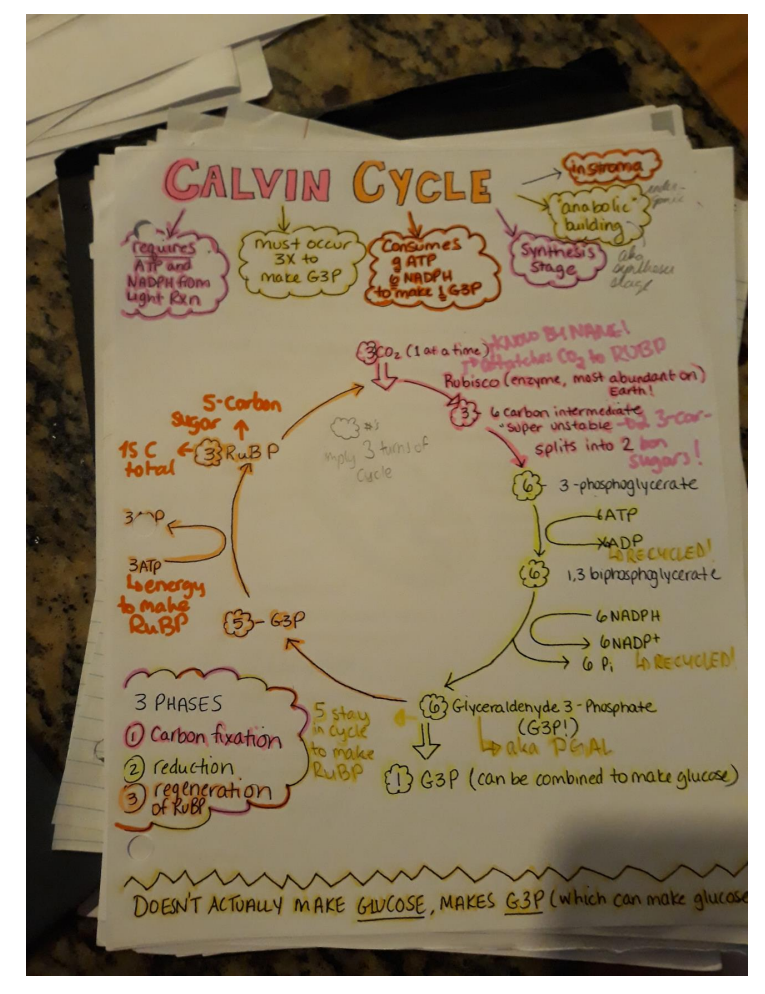
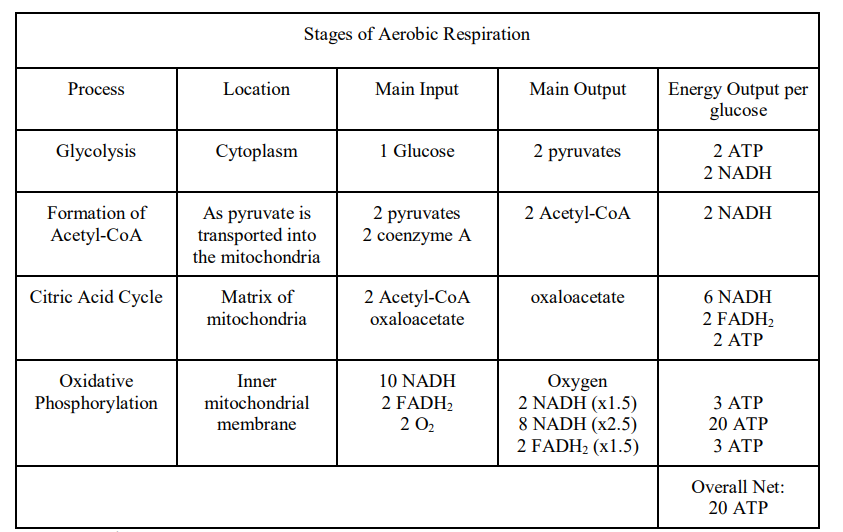
➢ 1. GLYCOLYSIS
- Glucose is split
- Glucose 6-carbon; when it is split it makes 2 3-carbon pyruvates
- Creates 2 ATP (net)
- NADH created from the transfer of electrons to NAD+
- Occurs in cytoplasm
- Glucose + 2 ATP + 2 NADh ⟶2 Pyruvate + 4 ATP + 2ND
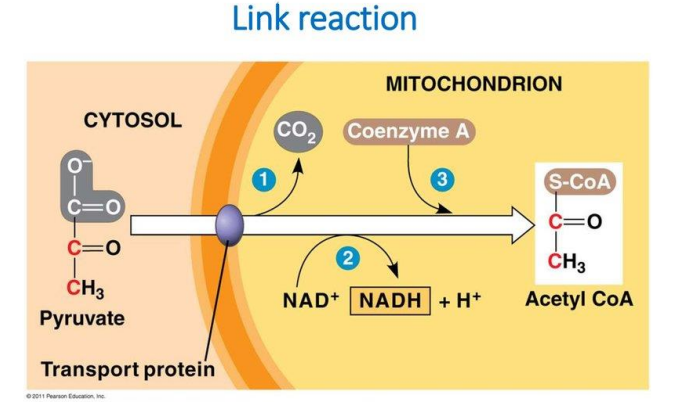
➢ 2. FORMATION OF ACETYL CoA
- $ 2Pyruvate + 2 Coenzyme A + 2 NAD^+ ⟶2 Acetyl-CoA + 2CO^2 + 2 NADH$
- Extra carbons leave cell as $CO_2$
➢ 3. CITRIC ACID CYCLE
- Aka Krebs cycle
- Each acetyl coa will enter Krebs cycle on at a time, and all carbons will be turned into $CO_2$ eventually
- Acetyl $CoA$ combines with oxaloacetate (4-carbon) to create citric acid
- Active transport into mitochondria via cotransport with oxygen
- citric eventually gets turned back into oxaloacetate
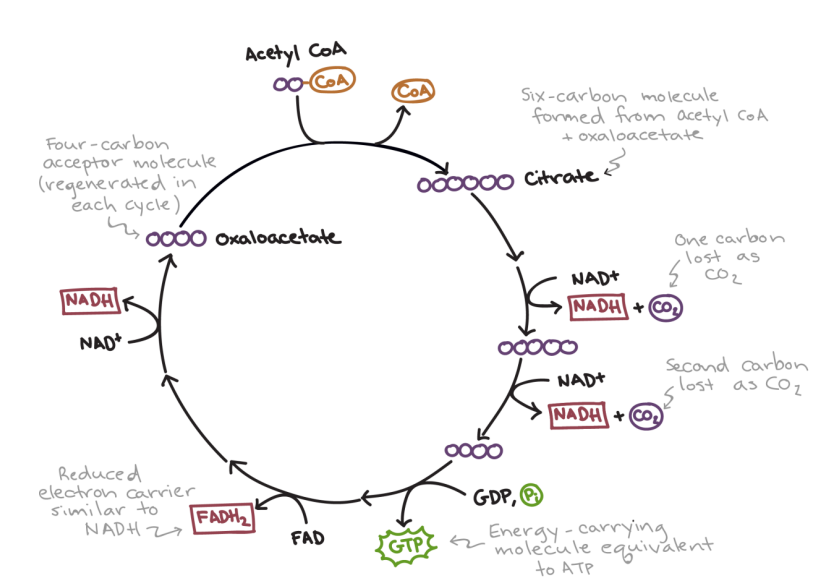
- 3 types of energy produced:
■ $1ATP$
■ $3 NADH$
■ $1 FADH_2$
○ Atthis point there are 4 ATP, 10 NADH, and 2 FADH2 total
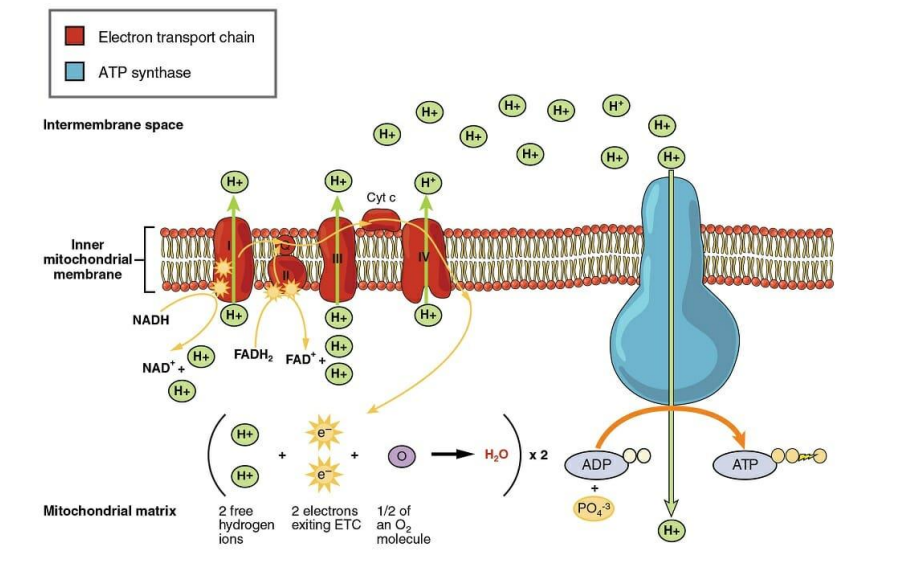
➢ 4. OXIDATIVE PHOSPHORYLATION
- As electrons are removed from a molecule of glucose, they carry with them as much of the energy that was originally stored within their bonds
- These electrons are then transferred to readied carrier molecules–NADH and $FADH_2$
- Electron carriers shuttle electrons down to electron transport chain, and the resulting $NAD^{+}$ and FADH can be recycled to be used again
■ Hydrogen atoms are split
● $\mathrm{H}_2-2 \mathrm{H}^{+}+2 e^{-}$
■ High-energy electrons are passed down a series of protein carrier molecules that are embedded in the cristae
● Some proteins include NADH dehydrogenase and cytochrome C
■ The electrons travel down the electron transport chain until they reach the final acceptor, oxygen
● Oxygen pulls the electrons through the chain due to its electronegativity and then combines with them and hydrogen to create water
● Allows For a gradual release of energy rather than a sudden, explosive one - Chemiosmosis
■ The energy released from the ETC is used to pump hydrogen ions across the inner mitochondrial membrane from the matrix into the intermembrane space
● Creates pH/proton gradient
● Potential energy created from gradient is responsible for the production of ATP - Flow back in through ATP synthase and this movement provides the energy necessary to produce ATP
- $ADP + P_i ⟶ATP$
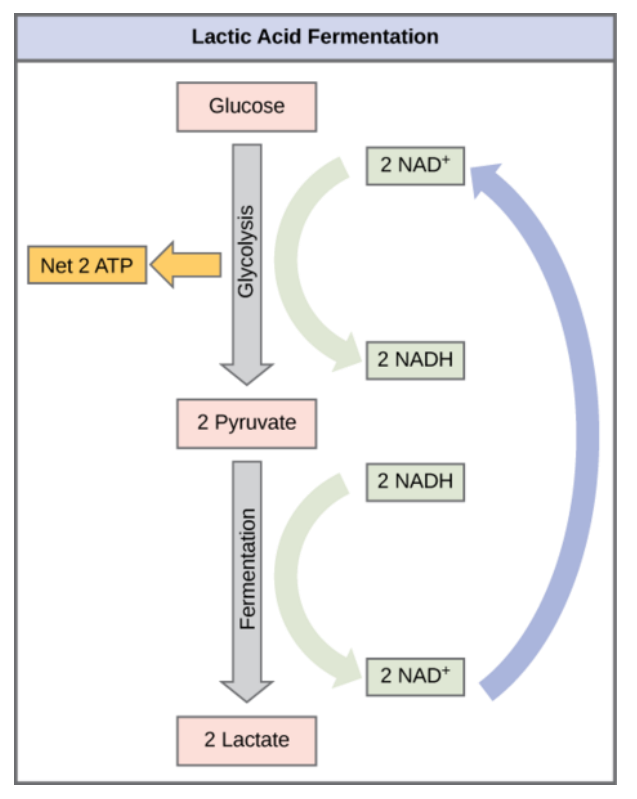
G, Fermentation
➢ In anaerobic environments, cellular respiration doesn’t work
- No ETC, so electron carriers are useless
- No Acetyl CoA or Citric Acid cycle
- Only glycolysis can occur
➢ Glycolysis produces 2 pyruvate and $\text{2 NADH}$
- In order to recycle NADH, pyruvate takes its electrons, creating lactic acid in muscles or ethanol in yeast
■ Both products are unfortunately toxic - ATTP created through substrate-level phosphorylation
➢ Common in bacteria
- In some, an ETC may exist, but $SO_4$ is the electron acceptor instead of $O_2$, creating $H_2SO_4$ as a byproduct