A.Molecular Structure of DNA
➢ Made up of repeated units of nucleotides
- Each nucleotide has:
■ 5-carbon sugar
● Pentagon shaped
● Called deoxyribose
● Linked to phosphate and nitrogenous base
■ Phosphate
■ Nitrogenous base
● Adenine - Purine (double-ringed)
● Guanine - Purine
● Cytosine - Pyrimidine (single-ringed)
● Thymine - Pyrimidine
● Purines always pair up with pyrimidines to keep DNA width consistent
➢ Nucleotides linked together by phosphodiester bonds that make up the sugar-phosphate backbone
➢ Double helix structure discovered by scientists Watson, Crick, and Franklin
- Nobel prize earned for discovery
- Franklin used X-Ray crystallography in its discovery
➢ Base pairing (Chargaff’s Rules)
- Each base can only bond with a specific complementary base
■ A-T
■ C-G - Specific rations of each nucleotides in the same species
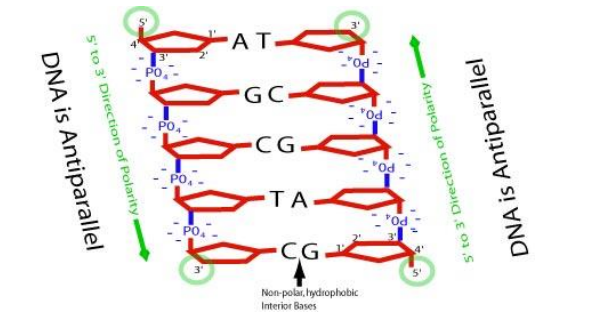
➢ Complementary strands
➢ Strands are antiparallel
- Run in opposite directions
- 5’ and 3’ end named after carbons that end them
■ 5’ has phosphate group
■ 3’ has hydroxyl group - Strands linked by hydrogen bonds
B. GEnome Structure
➢ Genetic code is the sequence of the base pairs
➢ Gene codes for a specific protein
➢ Prokaryotes: single DNA molecule
➢ Eukaryotes: multiple DNA molecules
➢ An species entire DNA sequence is its genome
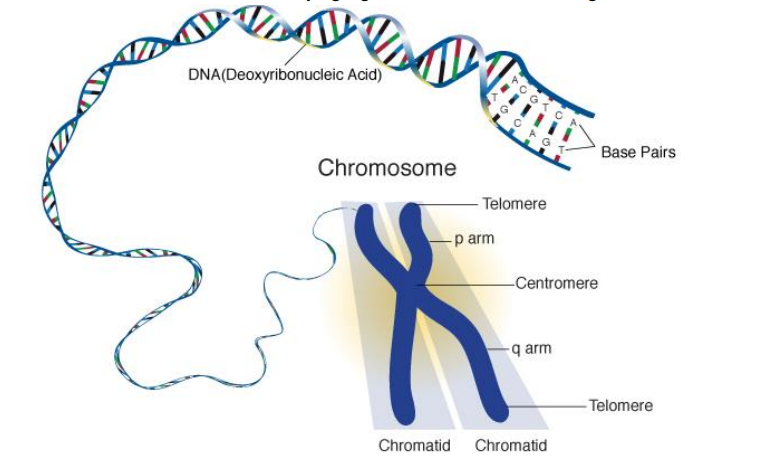
➢ Chromosome is each separate chunk of DNA
➢ Prokaryotes have one circular chromosome, and eukaryotes have linear chromosomes
➢ chromosomes wrapped around proteins called histones, and histones are bunched in groups called a nucleosome
➢ How Tightly DNA is packaged depend on the section of DNA and what is going on in the cell at the time
- Euchromatin is extremely loose genetic material
- Heterochromatic is extremely tight genetic material with inactive genes
C. DNA Replication
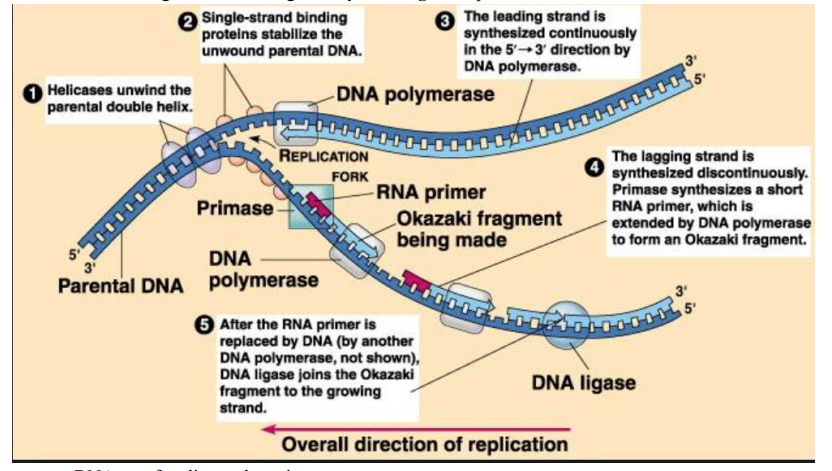
➢ First step is to unwind the double helix by breaking the hydrogen bonds
- Accomplished by an enzyme helicase
- Single stranded Binding Protein hold the strands apart
- Origin of replication=place where replication process begins; short stretch of DNA with specific nucleotide sequence
- Exposed DNA now forms a y-shaped replication fork
➢ DNA replication begins at specific sites called origins of replication
➢ Topoisomerase cuts and rejoins the helix to prevent tangling and relieve tension
➢ DNA polymerase III adds nucleotides to freshly built strand
- Can only add nucleotides to the 3’ end
➢ RNA primase adds a short strand of RNA nucleotides called an RNA primer
- Primase synthesizes RNA primer
- After replication, the DNA Polymerase I removes the RNA primer and replaces it with DNA
➢ Leading strand
- Synthesized continuously
- $5’ to 3’$
- Replicated towards fork
➢ Lagging strand
- Made in pieces called Okazaki fragments
- $3’ to 5’$
- Replicated Away from fork
- Must be made in pieces since nucleotides can only added to 3’ end
- Fragments linked together by DNA ligase to produce a continuous strand
➢ DNA proofreading and repair
- DNA polymerase in charge of repair synthesis
- Nuclease removes damage
- Ligase seals newly repaired strands
- Repair enzymes detect damage
➢ Replicates semiconservatively because each new molecule is comprised of ½ of the original strand
- Semiconservative model proved by Meselson and Stahl’s experiments
■ Created “heavy” template DNA using N15, measured weights of replicated DNA by looking at the layers that formed, semiconservative model was the only one that fit
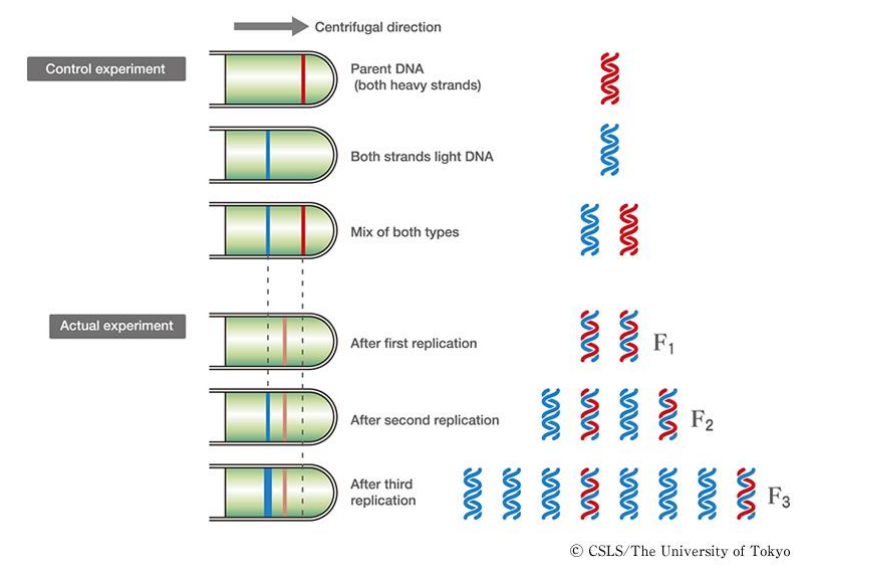
➢ A few bases at very end cannot be replicated because the DNA polymerase needs to bind
- Every time replication occurs the chromosome loses a few base pairs
- Genome has compensated for this over time by putting bits of unimportant/less important DNA at the ends of a molecules called telomere
➢ Key history
- Protein originally thought to be the carrier of genetic material due to its higher variety and specific functions
- Griffiths want to find out what substance causes transformation
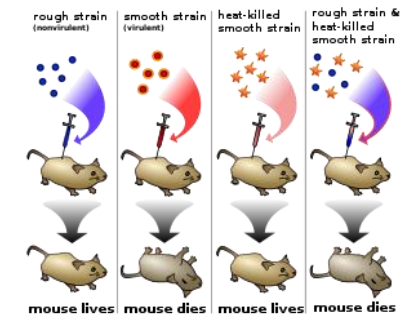
R bacteria had been transformed into pathogenic S bacteria by unknown substance
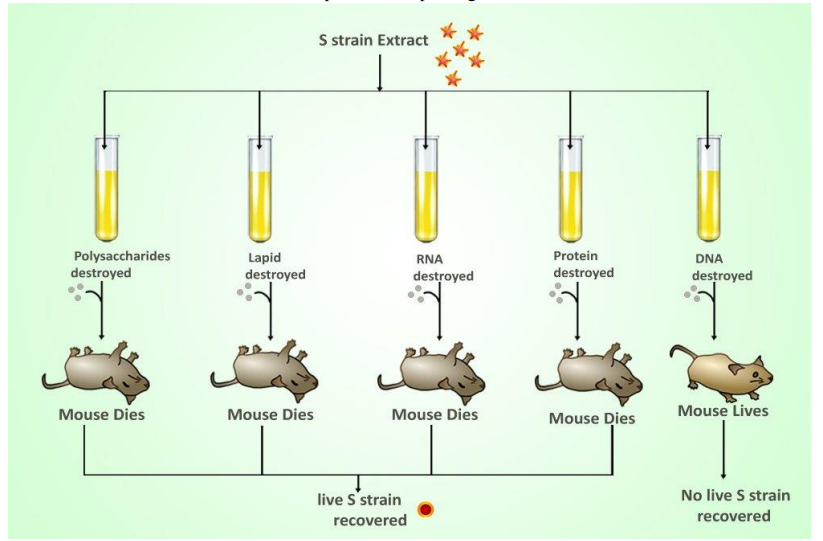
- A very, MacLeod, and McCarty isolated various cellular components from a dead virulent strain of bacteria
■ Followed up on a previous experiment by Griffiths and added each of these cellular components to a strain of living nonvirulent bacteria
■ Only the component of the deadly bacteria was able to change the second bacteria into a deadly strain capable of reproducing
■ DNA must be responsible for passing traits and it is inheritable
- Hershey-CHase
■ Used bacteriophages and labelled protein parts of some with radiolabeled sulfur and labelled the DNA parts of other viruses with radiolabeled phosphorus
■ Bacteriophages inject genetic material into cell so more genetic material will be created
■ When viruses infected bacteria, only the labelled DNA was inside, but they were still able to replicate and make progeny viruses
■ E. coli were infected by the phage, and there was more and more P that entered. They concluded that DNA carried the genetic information to produce DNA and proteins
➢ Central Dogma
- DNA’s main role is directing the manufacture of molecules that actually do the work in the body
- DNA expression:
- 1. Turn into RNA
- 2. Send RNA out into the cell and often gets turned into a protein
➢ Transcription turns RNA into DNA
- Takes place in nucleus (except in prokaryotes)
➢ Translation turns RNA into a protein
- Takes place in cytoplasm
➢ RNA
- Single stranded
- 5-carbon sugar is ribose instead of deoxyribose
- Uses uracil instead of thymine
- major types of RNA
■ Messenger RNA (mRNA)
● Temporary version of DNA that gets sent to ribosome
■ Ribosomal RNA (rRNA)
● Produced in nucleolus
● Makes up part of ribosomes
■ Transfer RNA (tRNA)
● Shuttles Amino acids to the ribosomes
● Responsible for bringing the appropriate amino acids into place at the appropriate time
● Done by reading message carried by mRNA
■ Interfering RNA (RNAi)
● Small snippets of RNA that are naturally made in the body or intentionally created by humans
● siRNA and miRNA can bind to specific sequences of RNA and mark them for destruction
➢ Transcription
- RNA copy of DNA code
■ pre-mRNA synthesis - Only a specific section is copied into mRNA
- Occurs as-needed on a gene-by-gene basis
- Exception: prokaryotes will transcribe a recipe that can be used to make several proteins
■ Called polycistronic transcript
■ Eukaryotes tend to have one gene that gets transcribed to one mRNA and translated into one protein
● Monocistronic transcript
- 3 steps: initiation, elongation, termination
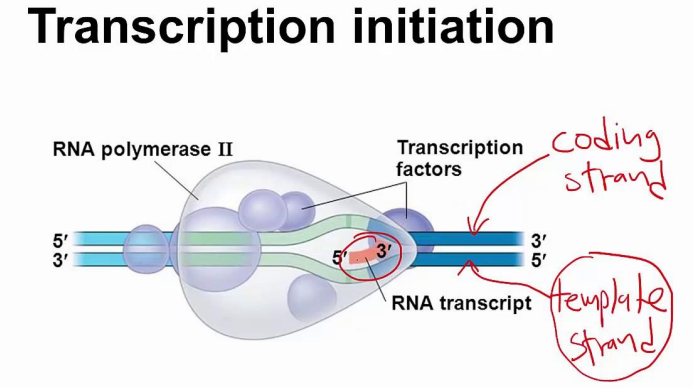
■ initiation
● Unwind and unzip DNA strands using helicase
● Transcription initiation complex - Transcription factor proteins+RNA polymerase
- Forms at promoter
● Transcription only occurs as-needed to conserve resources
■ ELongation
● Begins at special sequences of the DNA strand called promoters
● Free RNA nucleotides inside the nucleus used to create mRNA - RNA polymerase used to construct mRNA
● Strand that serves as the template is called antisense strand, the noncoding strand, or the template strand
● Strand that lies dormant is the sense strand, or the coding strand
● Rna polymerase build RNA only to 3’ side - Doesn’t need primer
● Promoter region is “upstream” of the actual coding part of the gene
● Official starting point if start site
● RNA strand is complementary to template DNA strand
■ Termination
● Once termination sequence is reached, it separates from the DNA template, completing the process of transcription
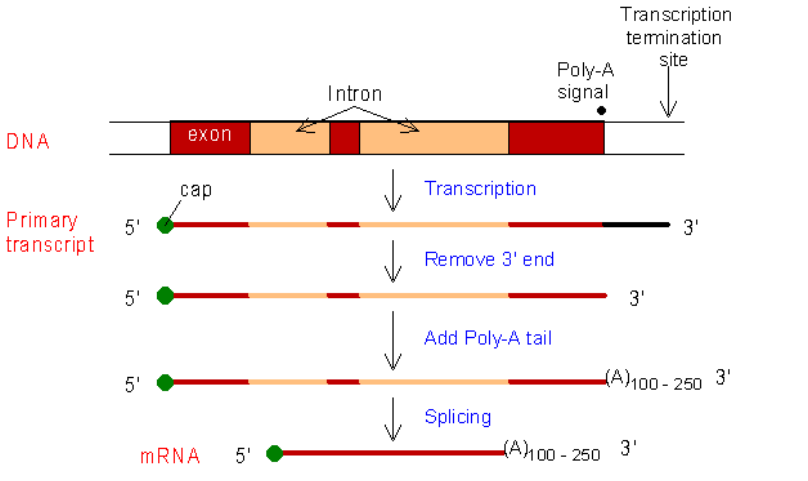
➢ RNA processing
- In eukaryotes the RNA must be processed before it can leave the nucleus
- Freshly transcribed RNA is called hnRNA (heterogenous nuclear RNA) and it contains both coding regions and noncoding regions
- Regions that express the code will be turned into protein are exons
- Non-coding regions in the mRNA are introns
■ Introns removed by spliceosome
● Spliceosome made up of many snRNPs - snRNPs made up of ribozyme+small nuclear RNA
■ ribozyme=RNA catalyst that can copy RNA strands
● Spliceosome identifies ends of an intron
● Folds chromosome
● Spliceosome cuts out the intron and binds the two exons together
● Non i prokaryotic cells
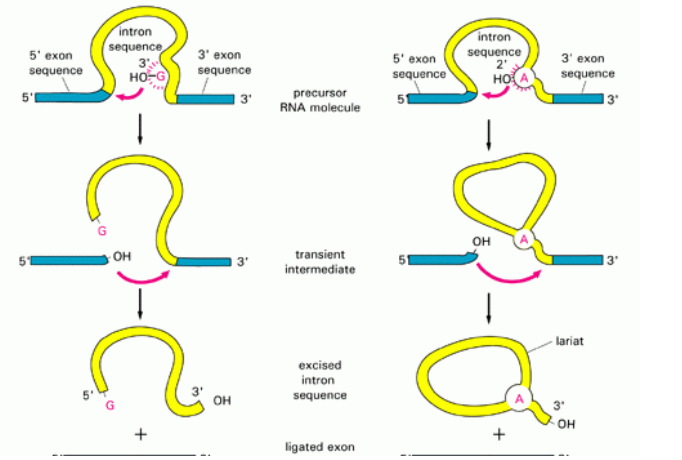
● 3 properties of RNA that allow it to function as an enzyme
- Single stranded
- Functional groups that act as catalysts
- Hydrogen bonds with other nucleic acids
■ Introns allow more genetic diversity
● More possibilities for crossover
● Alternate splicing can yield new protein varieties - Methyl cap added to 5’ end
■ Helps mRNA leave nucleus
■ Allows attachment to ribosome
■ Modified guanine
○ poly-A tail added to 3’ end
■ Protects mRNA from endonucleases in cytoplasm, which can only attach to 3’ end
■ 50-250 adenine
○ No cap or tail in prokaryotic cells since they have no nucleus
○ mRNA leaves nucleus through nuclear pore
➢ TRanslation
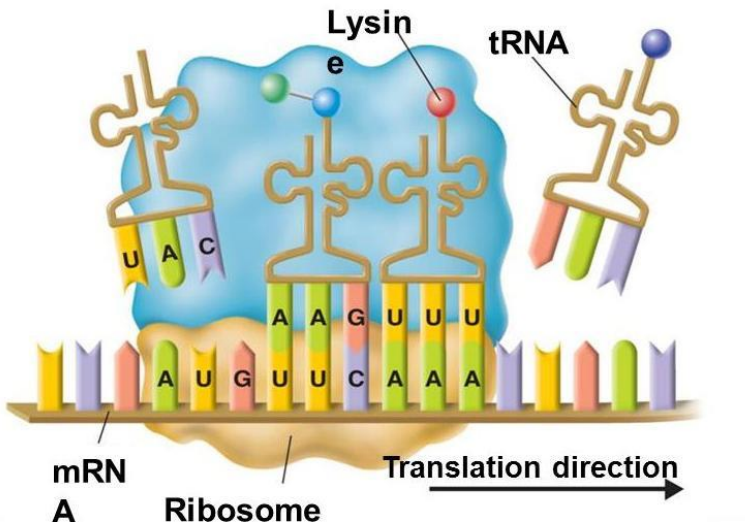
- Process of turning mRNA into a protein
- mRNA nucleotides will be read in the ribosome in groups of three
■ Group of three nucleotides=codon
● Each codon corresponds to a particular amino acid - mRNA attaches to the ribosomes to initiate translation and “waits” for the amino acids to come to the ribosome
- 3 sites on ribosome
■ E=exit site
■ P=polypeptide storage/exit
■ A= place where tRNA brings in amino acid - 1.mRNA attaches to mRNA binding site on small subunit. tRNA attaches onto A site
- 2. Large subunit attaches via GTP
■ 1st tRNA is in P site
■ 2nd tRNA comes in A site - 3. rRNA in large subunit catalyzes a peptide bond between amino acids
○ 4. 1st rRNA moves to exit site
■ 2nd tRNA moves to P site
■ New tRNA comes in through A site
■ Steps 1-4 repeats until stop codon is reached
■ As mRNA moves through ribosome, other ribosomes can attach to it at the same time (as long as mRNA has not degraded, especially on 5’cap
- 5. Release factor adds water to end of polypeptide; polypeptide detaches and exits
through P-site tunnel - 6. small/large subunit/mRNA disassemble and disassociate; process of translation can start over again
- tRNA carries amino acid
■ Attaches to RNA via anticodon (complementary base pair to codon)
■ Wobble pairing on third nucleotide (flexible bonds)
■ Each tRNA becomes charged and enzymatically attaches to an amino acid in the cell’s cytoplasm and “shuttle” it to the ribosome
■ Charging enzymes require ATP - 3 phases:
■ Initiation
● 3 binding sites: - A site
- P site
- E site
● Start codon is AUG (methionine)
● TATA box=specific promoter for initiation
● RNA polymerase binds to a specific location on promoter
● Transcription factors attach to promoter to help guide RNA polymerase
■ Elongation
● As each amino acid is brought to the mRNA, it is linked to its neighboring amino acid by a peptide bond and eventually forms a full protein
■ Termination
● Synthesis of a polypeptide ended by stop codons
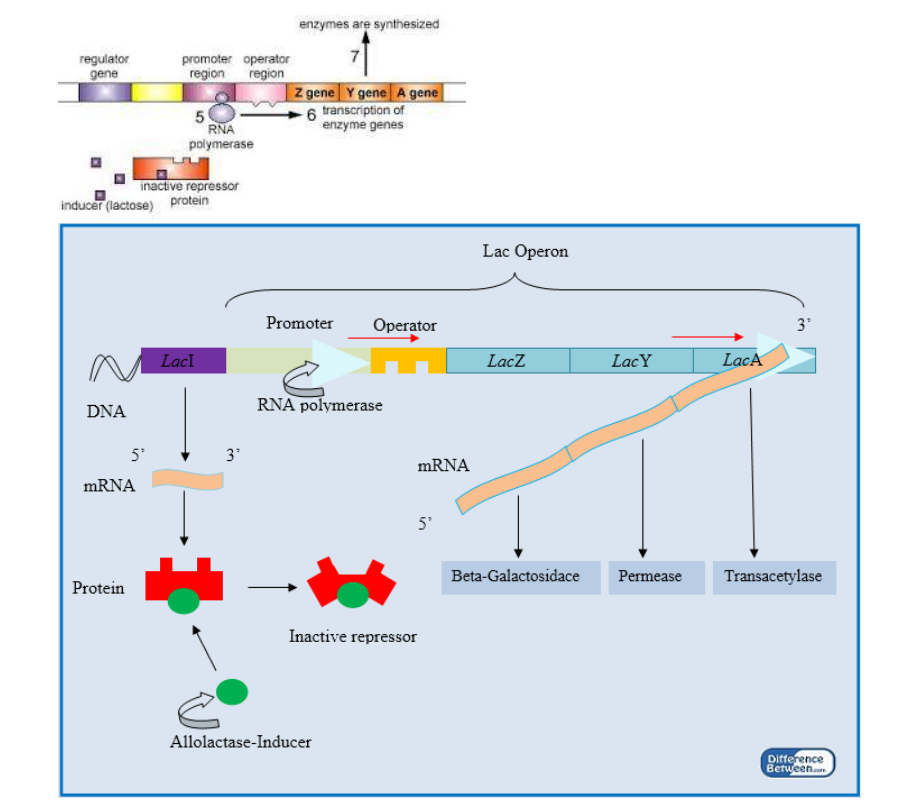
➢ Gene Regulation
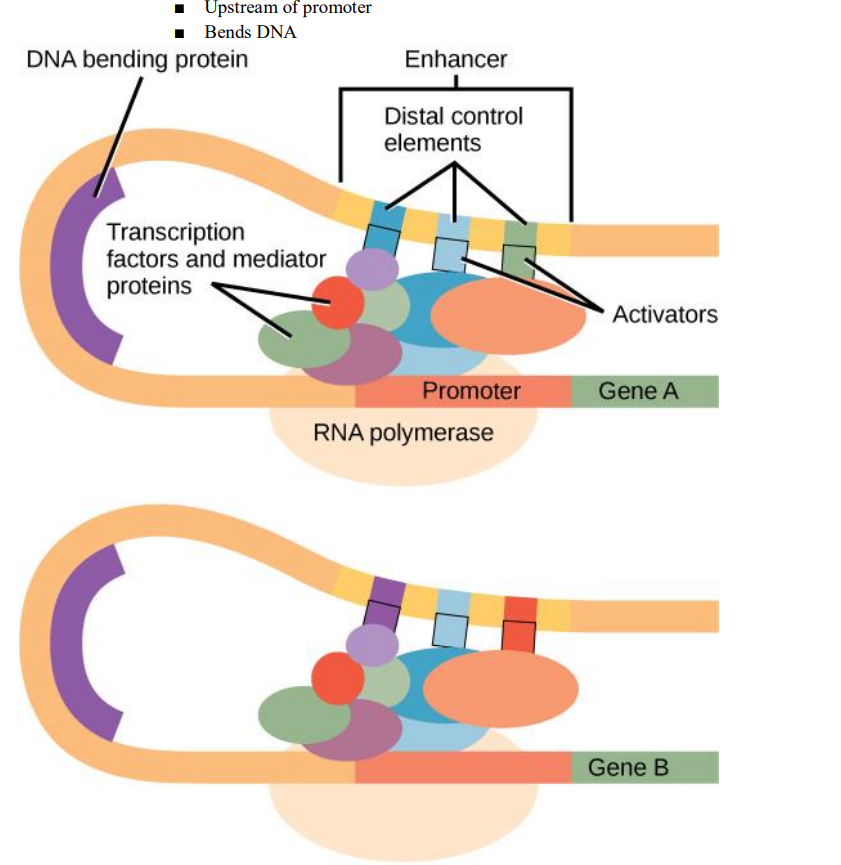
- Pre-transcriptional regulation
■ Transcription factors can either encourage or inhibit the unwinding of DNA and the binding of RNA polymerase - Operons
■ Bacteria only
■ Structural genes
● Code for enzymes in a chemical reaction
● Genes will be transcribed at the same time to produce particular enzymes
■ Promoter gene
● Region where RNA polymerase binds to begin transcription
■ Operator
● Controls whether transcription will occur
● Where repressor/inducer binds
■ Regulatory gene
● Codes for a specific regulatory protein called to repressor
● Repressor capable of attaching to the operator and blocking transcription
● If repressor binds to the operator, transcription will not occur
● If repressor does not bind to the operator, RNA moves along operator and transcription occurs
■ inducible=presence of molecule turns gene on
■ repressible=presence of molecule turns gene off
- Chromatin Modification
■ Histone acetylation
● Acetyl groups added to histones
● Looser Chromatin
● Access for transcription increased
■ DNA methylation
● Methyl groups added to bases
● Silences gene
● Tightens chromatin - Enhancers
- Post-transcriptional regulation
■ Occurs when the cell creates an RNA but then decides that it should not be translated into a protein
■ RNAi molecules bind to an RNA via complementary base pairing
■ Creates double-stranded RNA, signalling that RNA should be destroyed, preventing it from being translated - Post-translational regulation
■ Protein has already been made, but doesn’t need it yet, so it is deactivated
➢ Mutations
- Mutation is an error in the genetic code
- Occur because DNA is damaged in cannot be repaired or because DNA is repaired incorrectly
■ Caused by chemicals or radiation
■ Can also occur when a DNA polymerase or an RNA polymerase makes a mistake
■ RNA polymerases have proofreading abilities, but RNA polymerases do not - Error in DNA not a problem unless that gene is expressed AND the error causes a change in the gene product
➢ Base Substitution
- Point mutations result when a single nucleotide is replaced for another
- Nonsense mutation
■ Cause original codon to become a stop codon, which results in early termination of protein synthesis - Missense mutation
- Cause original codon to be altered and produce a different amino acid
- Silent mutations
■ codon that codes for the same amino acid is created and therefore does not
change the corresponding protein sequence - Frameshift mutations
■ insertions/deletions result in the gain or loss of DNA or a gene
● Can have devastating consequences during translation - results in a change in the sequence of codons used by the ribosome
- Duplications
■ Extra copy of genes
■ Caused by unequal crossing-over during by meiosis or chromosome
rearrangements
■ ,ay result in new traits as one copy may evolve a new function
○ Inversions
■ Changes occur in the orientation of chromosomal regions
■ May cause harmful effects if the inversion involves a gene or an important regulatory sequence - Translocation
■ 2 different chromosomes break and rejoin in a way that causes the DNA sequence to be lost, repeated, or interrupted
D. Biotechnology
➢ Recombinant DNA generated by combining DNA from multiple sources to create a unique DNA molecule that is not found in nature
○ Ex. introduction of a eukaryotic gene of interest into a bacterium for production
➢ Polymerase Chain Reaction (PCR)
- Enables the creation of billions of copies of genes within a few hours
- DNA polymerase, DNA, and lots of nucleotides added in a small PCR tube
- Thermocycler
■ PCR machine that heats, cools, and warms PCR tubes many times
■ Each time the machine is heated, the hydrogen bonds break, separating the double-stranded DNA (Denaturation)
■ As it cools, primers bind to the sequence flanking the region of the DNA we want to copy, primers can form hydrogen bonds with ends of target sequence (Annealing)
■ When it is warmed, polymerase binds to the primers on each strand and adds
nucleotides on each template strands (extension)
■ REPEAT exponentially
➢ Transformation
- Transformation: process of giving bacteria foreign DNA
■ Genes of interest (vectors) placed into small circular DNA molecule called a plasmid
● Plasmid usually codes for antibiotic resistance
● Small ring of DNA found in bacteria that is replicated separately form the chromosomal DNA
● Not in all bacteria
■ 1. Extract the plasmid
■ 2. Add a restriction enzyme that will cut the ring open
● Restriction enzymes usually used to cut up foreign DNA, but are used by
scientists for this purpose
● Cuts palindromes, leaving “sticky ends”
● Always cut at the same nucleotide sequence
■ 3. Cut a piece of human DNA with same restriction enzyme
● Reverse transcriptase must be used to process DNA since prokaryotes do not have RNA processing to remove introns - DNA transcribed and mRNA is processed, and then reverse transcriptase turns mRNA back into DNA
● Reverse transcriptase also used by retroviruses
● Problems: vector may be too big, and there is no direct way to force the plasmid to accept the vector
■ 4. Mix the cut plasmids with the cut human DNA-some will align right due to their sticky ends
● Ligase used to glue ends back together
■ 5. Allow bacteria to take plasmid back in
● Heat shock/electric shock used to change membrane so plasmid can reenter easily
■ 6. Allow to reproduce
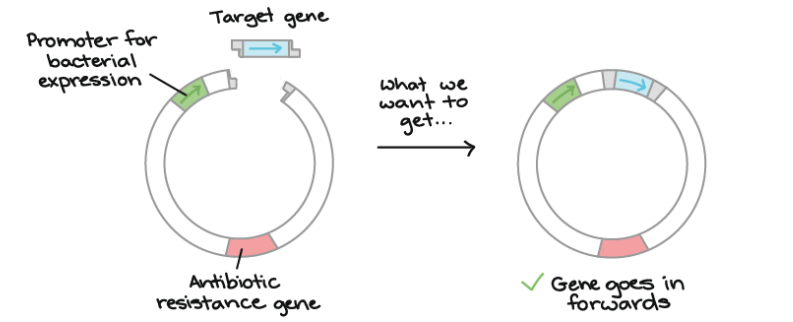
■ Not all bacteria will be transformed, can be tested by using antibiotic resistance
■ Allows the safe mass-production of proteins used for medicine
■ Important role in the study of gene expression
■ Transfection: putting a plasmid into a eukaryotic cell, rather than a bacteria cell
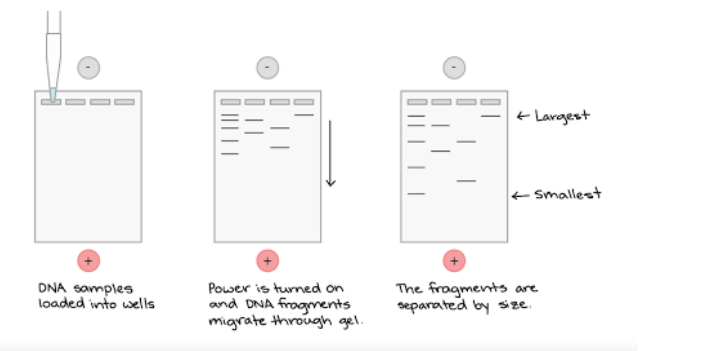
➢ Gel Electrophoresis
- DNA fragments can be separated according to their molecular weight using gel electrophoresis
- DNA put into wells on negative end, and when a current is run through the gel, the DNA moves across gel according to their weight
- Because DNA and RNA are negatively charged, they migrate through the gel toward the positive pole of the electrical field
■ Smaller fragments move faster and farther - Restriction enzymes used to create a molecular fingerprint
■ Places where enzymes cut and thus the sizes are unique for each person
➢ Stem cells also very important in biotechnology since they can turn into many different kinds of cells, but it is controversial due to harvesting methods
- Totipotent cell: capable of giving rise to any type of cell or a complete embryo
- Pluripotent cell:capable of giving rise to different cell types
➢ Cell division
- Mechanism to replace dying cells
- Small part of life cycle of a cell
- Some types of cells are nondividing
■ Usually highly specialized cells derived from a less specialized type of cell
■ Made as needed, but cannot replicate themselves
■ Ex. red blood cells - Multicellular organisms depend on cell division for:
■ Development from a fertilized cell
■ Growth
■ Repair
➢ Binary Fission
- Used by prokaryotes
- Chromosome replicates at origin of replication and the two daughter chromosomes actively move apart
- Plasma membrane pinches inward, dividing cell into two Mitosis likely evolved from binary fission
■ Certain protists exhibit cell division that seem intermediate between binary fission and mitosis
A. Interphase
➢ Time span from one cell division to another
➢ Cell carries out regular activities
➢ All the proteins/enzymes the cell needs to grow are produced in interphase
➢ 3 Phases:$G_1$, S,$G_2$
- $G_1$
■ Cell produces all enzymes required for replication
■ G=”Gap” or “growth” - S
■ Cell replicates genetic material
■ Every chromosome in nucleus is duplicated
● Sister chromatids created, held together by centromere
■ To be called a chromosome, they each need to have their own centromere; once chromatids separate, they will be “chromosomes”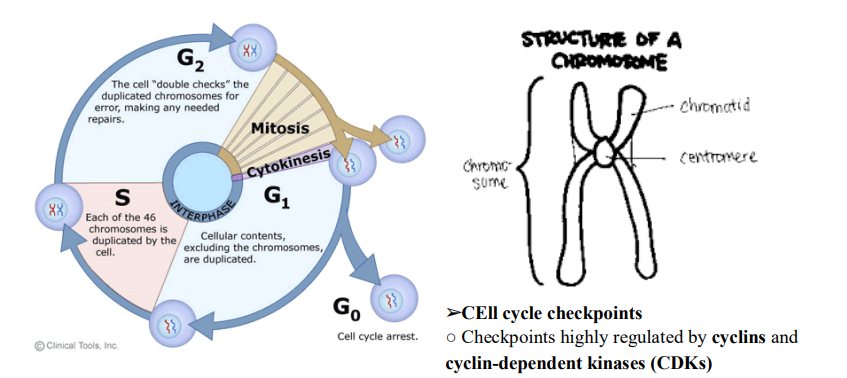
■ To induce cell cycle progression, CDK binds to a regulatory cyclin. Once together, the complex is activate
● Can affect many proteins in cell
● Causes cell cycle to continue
● To inhibit cell cycle progression, CDKs and cyclins are kept separate
● Separated via dephosphorylation
■ MEtaphase Checkpoint
● Chromosome spindle attcachment
■ $G_1$ Checkpoint
● Check for:
- Nutrients
- Growth factors
- DNA damage
● Can put cell into $G_0$ is it doesn’t need to divide
■ $G_2$ checkpoint
● Check for: - Cell size
- DNA replication
- Make sure cell division is happening properly in cells
- Stops progression if cell is not ready to progress to next stage
- In eukaryotes, checkpoint pathways mainly function ay phase boundaries
- When DNA damage is detected, cell will not progress until damage is fixed, or apoptosis is started
- Cancer can result from a mutation in a protein that normally controls progression, resulting in unregulated cell division
■ Oncogenes are genes that cause cancer
● Normally required for proper growth and regulation of he cell cycle
● Mutated versions can cause cancer
● proto-oncogene=normal, healthy oncogene
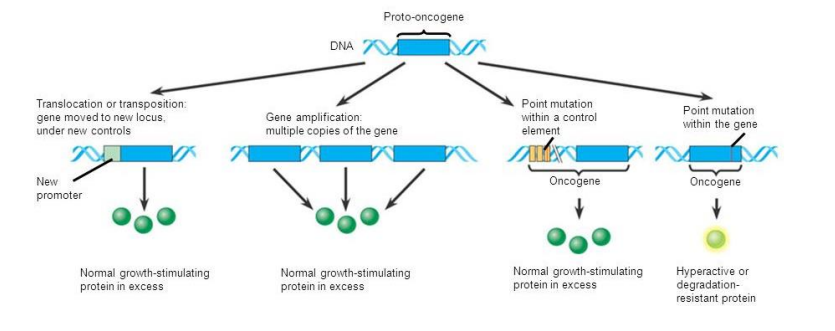
■ Tumor suppressor genes
● Produce proteins that prevent the conversion of normal cells into cancer cells
● Detect damage within cell and work with CDK/cyclin complexes to stop cell growth until damage can be repaired
● Can trigger apoptosis is damage is too severe to be repaired
■ In order for a cell to become a cancer cell. It must simultaneously override checkpoints, grow in an unregulated way, and avoid cell death
➢ Stop cell division
- Density-dependent inhibition
- Anchorage dependency
B. Mitosis
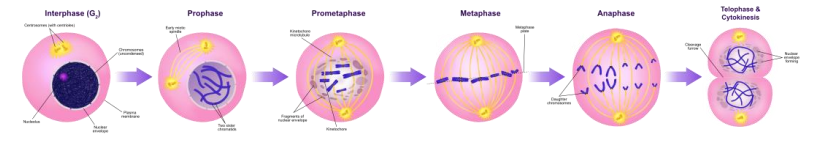
➢ Prophase
- Disappearance of the nucleolus and nuclear envelope
- Chromosomes thicken and become visible
■ Now called chromatin - Centrioles in microtubules organizing centers (MTOCs) start to move away from each other towards opposite poles of the cell
■ Centrioles spin out system of microtubules known as spindle fibers
■ Spindle fibers attach to kinetochore located on centromere of each chromatid
➢ Metaphase
- Chromosomes begin to line up along equatorial metaphase plate
■ Moved along by spindle fibers attach to kinetochores on each chromatid
➢ Anaphase
- Sister chromatids of each chromosome separate at the centromere and migrate to opposite
poles - Pulled apart by shortening microtubules
- Non-kinetochore tubules elongate cell
➢ Telophase
- Nuclear membrane forms around each set of chromosomes
- Nucleoli reappear
- Cytokinesis
■ Cytoplasm splits in half
■ Cell splits along cleavage furrow
■ Cell membrane forms along each new cell, split into distinct daughter cells
■ In plant cells, a cell plate forms down the middle instead of a cleavage furrow
➢ Interphase
- Cells re enter initial phase, and are ready to start the cycle over again
- Chromosomes become invisible again
■ Genetic material goes back to being chromatin
➢ Purpose of mitosis
- Produce daughter cells that are identical copies of parent cell
- Maintain proper number of chromosomes from generation to generation
➢ Occurs in almost every cell except for sex cells
➢ Involved in growth, repair, and asexual reproduction
C. Haploid vs. Diploid
➢ Diploid cell has 2 sets of chromosomes
- Most eukaryotic cells have 2 full sets of chromosomes: one for each parent
- Shown by “2n”
➢ Haploid cell has only one set of chromosomes
- Shown by “n”
➢ Homologous chromosomes are duplicate versions of each chromosome
- Similar in size and shape
- Express same traits, but may have different alleles
➢ Gametes
- Sex cells
- Haploid
■ Offspring will get one gamete from each parent, creating a diploid zygote/offspring
D. Meiosis
➢ Production of gametes
➢ Limited to sex cells in gonads
- gonads=sex organs
- Testes in males and ovaries in females
- Made up of germ cells
➢ Produces haploid cells which then combine to restore the diploid (2n) number during fertilization
➢ 2 rounds of cell division: meiosis I and meiosis II
➢ Just like in mitosis, double-stranded chromosomes are formed during S phase of interphase
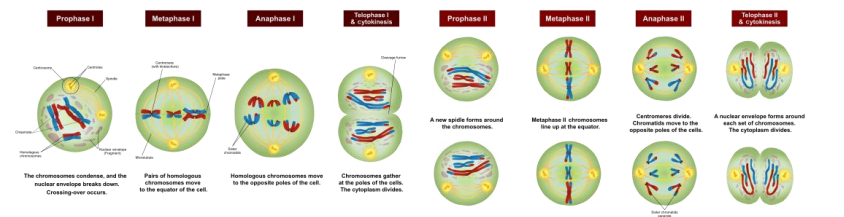
➢ Meiosis I
Prophase I
■ Nuclear membrane disappears
■ Chromosomes becomes visible
■ Centrioles move towards opposite ends of cell
■ Synapsis
● Chromosomes line up side-by-side with their homologs (counterparts)
● 2 sets of chromosomes come together to form a tetrad (aka bivalent) consisting of 4 chromatids
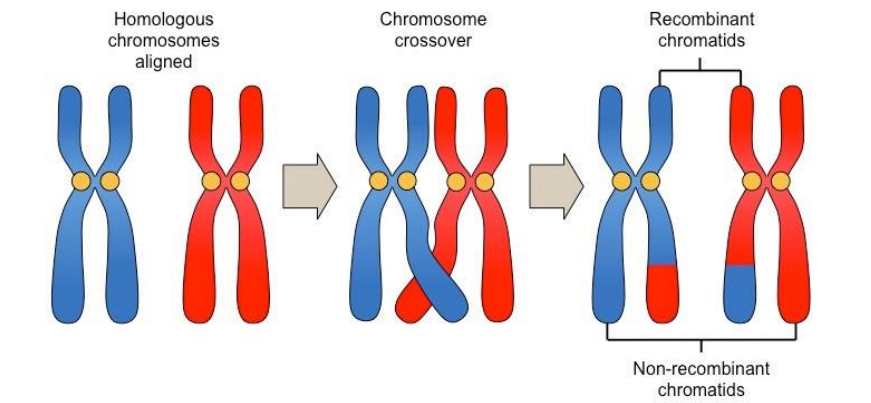
■ Crossing over
● Exchange of segments between homologous chromosomes
● Genetic variation
● Begins in Prophase I as homologous chromosomes line up gene by gene
● Produces recombinant chromosomes (DNA combined from each parent)
● Homologous portions of two nonsister chromatids trade placed
● Chromatids that are farther apart are more likely to cross over
- Metaphase I
■ Tetrads line up along metaphase plate
■ Random alignment–more genetic variation
● Offspring will be a combination of all 4 grandparents - ANaphase I
■ Each pair of chromatids within a tetrad separates and moves to opposite poles
■ Chromatids DO NOT separate at centromere - Telophase I
■ Nuclear membrane forms around each set of chromosomes
■ 2 daughter cells
■ Nucleus contains haploid number of chromosomes, but each chromosome is a duplicated chromosome consisting of 2 chromatids
➢ Meiosis II
- Purpose is just to separate sister chromatids
- Prophase II is the same
- Metaphase II: chromosomes move toward metaphase plate lining up in a single file, not in pairs
- Anaphase II:chromatids split at the centromere and each chromatid is pulled to opposite ends of cell
- Telophase II: nuclear membrane forms around each set of chromosomes and a total of 4 haploid cells are produced
- Meiosis I separates homologous chromosomes; Meiosis II separates sister chromatids
➢ Gametogenesis
- Spermatogenesis if sperm cells are produced
- Oogenesis if egg cell/ovum is produced
■ Produces one ovum instead of 4
■ Other 3 cells, called polar bodies get only a tiny amount of cytoplasm and eventually degenerate
■ Allows female to conserve as much cytoplasm as possible for the surviving ovum
➢ Meiotic Errors
- Nondisjunction: chromosomes fail to separate properly
■ Produces wrong number of chromones
■ Usually results in miscarriage or significant genetic defects
■ Ex. Down syndrome is a result of 3 copies of the 21st chromosome
- Translocation
■ One or more segments of a chromosome break and are either lost or reattach to
another chromosome
- Translocation