Question
Many biological processes are cyclical. Examples of cycles can be found at the level of the cell, the organism and the ecosystem.
(a) Explain how changes to the cell cycle can result in tumour formation.
(b) Outline the role of the right atrium in the cardiac cycle.
(c) Describe processes in the carbon cycle that produce or use carbon dioxide.
Answer/Explanation
Answer:
(a)
a. cell cycle is (repeated) sequence of cytokinesis/cell division, (then) interphase
(then) mitosis / cell cycle includes the sequence of interphase, mitosis and
cytokinesis/cell division (to form new cells which repeat the cycle);
b. cyclins control/regulate this cycle / ensure the cell moves on to the next stage
of the cycle when it is appropriate;
c. extra cells produced when they are needed;
d. tumour formation is the result of uncontrolled cell division/ cells growing and
dividing endlessly;
e. repeated mitoses/high mitotic index;
f. due to mutations in oncogenes / oncogenes may become active and contribute
to the development of a cancer cell;
g. carcinogens/radiation/mutagenic chemicals/mutagens/smoking cause
mutations/tumours;
(b)
a. right atrium collects (deoxygenated) blood from the body;
b. blood drains into atrium through/from the vena cava;
c. deoxygenated blood present in vena cava/right atrium/right ventricle;
d. right atrium pumps blood into the right ventricle;
e. during atrial systole/ventricular diastole/at the start of the cardiac cycle;
f. sinoatrial node is in the right atrium/sinoatrial node acts as the (natural)
pacemaker / sinoatrial node initiates the heartbeat;
g. SA node sends out electrical signal to stimulate contraction in the (walls of the)
atria/then propagated to the AV node / (walls of the) ventricles;
(c)
a. photosynthesis uses carbon dioxide / reduces carbon dioxide concentration of the atmosphere;
b. autotrophs/plants/cyanobacteria convert/fix carbon dioxide into carbon/organic
compounds;
c. cell respiration produces/releases carbon dioxide;
d. glucose/carbon/organic compounds oxidised/broken down to produce/release
carbon dioxide;
e. carbon dioxide released from aerobic (cell) respiration AND anaerobic
respiration in yeast/plants (but not animals);
f. carbon dioxide released from saprotrophs/detritivores/decomposers from dead
organic matter / during decay/decomposition/respiration;
g. (partially) decomposed organic matter can lead to the formation of peat /
fossilized organic matter (coal/oil/natural gas)
h. carbon dioxide released when carbon/organic compounds burn / during
combustion (of biomass/fossil fuels) / forest fires;
i. carbon dioxide dissolves in aquatic ecosystems / can form carbonic
acid/hydrogen carbonate ions;
j. reef-building corals/molluscs use calcium carbonate to make/build
shells/exoskeletons or other body parts;
k. hard parts/shells/exoskeletons / precipitation of calcium carbonate to form
limestone/tufa;
Question
The diagram shows a section through the melatonin receptor, with melatonin attached to its binding site. Darker grey areas show the surface of the protein and paler areas are internal. The membrane in which this receptor is located is also shown.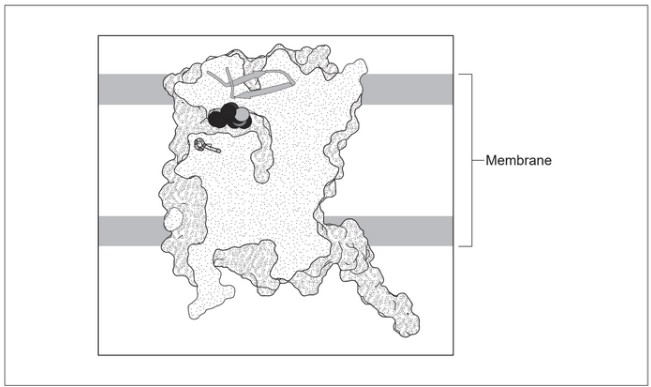
(a) Draw one phospholipid molecule on the diagram to show a possible position in the membrane.
(b) The receptor contains seven alpha helices and one other secondary structure.
Deduce what this other secondary structure is.
(c) Discuss briefly whether amino acids on the surface of the protein are likely to be polar or non-polar.
(d) Outline the role of melatonin in humans.
Answer/Explanation
Answer:
(a) phospholipid shown with circular head and 2 tails;
e.g.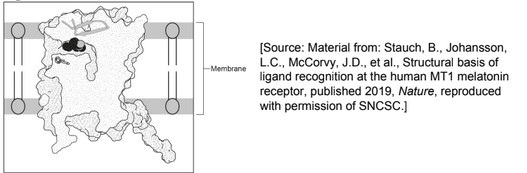
phospholipid shown in either the upper or lower half of the membrane only with its
head at the margin of the membrane and tails towards centre;
(b) beta pleated sheet/beta-loop/beta strands;
(c) polar/hydrophilic where exposed to the cytoplasm/to fluid outside cell/to polar phospholipid heads;
non-polar/hydrophobic where exposed to the (core of the) membrane/hydrophobic tails (of phospholipids);
(d)
a. control/maintain/regulate circadian rhythms;
b. secreted/released late evening/end of day/in dark/night time/dim light/absence of blue light;
c. helps to induce sleep/sleepiness/influences timing of sleeping/waking/control sleep cycle;
Question
(a) Outline the functions of rough endoplasmic reticulum and Golgi apparatus. [3]
(b) Outline the control of metabolism by end-product inhibition. [5]
(c) Explain how hydrophobic and hydrophilic properties contribute to the arrangement of molecules in a membrane. [7]
▶️Answer/Explanation
Ans:
a
a.ribosomes on RER synthesize/produce polypeptides/proteins ✔
b.proteins from RER for secretion/export/use outside cell/for lysosomes ✔
c.Golgi alters/modifies proteins/example of modification ✔
d.vesicles budded off Golgi transport proteins «to plasma membrane»
OR
exocytosis/secretion of proteins in vesicles from the Golgi ✔
b
a.metabolism is chains/web of enzyme-catalyzed reactions
OR
metabolic pathway is a chain of enzyme-catalyzed reactions ✔
b.end product/inhibitor is final product of chain/pathway ✔
c.inhibits/binds to/blocks the first enzyme in chain/pathway ✔
d.non-competitive inhibition ✔
e.end-product/inhibitor binds to an allosteric site/site away from the active site ✔
f.changes the shape of the active site/affinity of the active site «for the substrate» ✔
g.prevents intermediates from building up
OR
prevents formation of excess «end» product/stops production when there is enough
OR
whole metabolic pathway can be switched off ✔
h.negative feedback ✔
i.binding of the end product/inhibitor is reversible
OR
pathway restarts if end product/inhibitor detaches/if end product concentration is low ✔
j.isoleucine inhibits/slows «activity of first enzyme in» threonine to isoleucine pathway ✔
c
a. hydrophilic is attracted to/soluble in water and hydrophobic not attracted/insoluble ✔
b. hydrophilic phosphate/head and hydrophobic hydrocarbon/tail in phospholipids ✔
c. phospholipid bilayer in water/in membranes ✔
d. hydrophilic heads «of phospholipids» face outwards/are on surface ✔
e. hydrophobic tails «of phospholipids» face inwards/are inside/are in core ✔
f. cholesterol is «mainly» hydrophobic/amphipathic so is located among phospholipids/in hydrophobic region of membrane ✔
g. some amino acids are hydrophilic and some are hydrophobic ✔
h. hydrophobic «amino acids/regions of» proteins in phospholipid bilayer «core» ✔
i. hydrophilic «amino acids/regions of» proteins are on the membrane surface ✔
j. integral proteins are embedded in membranes due to hydrophobic properties/region OR
transmembrane proteins have a hydrophobic middle region and hydrophilic ends ✔
k. peripheral proteins on are on the membrane surface/among phosphate heads due to being «entirely» hydrophilic
OR
«carbohydrate» part of glycoproteins is hydrophilic so is outside the membrane ✔
l. pore of channel proteins is hydrophilic ✔
Question
Proteins are an important group of chemicals found in all living organisms.
(a) Draw a molecular diagram to show the formation of a peptide bond. [3]
(b) Outline how proteins are digested and the products of protein digestion absorbed in humans. [4]
(c) Explain how polypeptides are produced by the process of translation. [8]
▶️Answer/Explanation
Ans:
a.
a two amino acids correctly drawn;
b removal/production of H2O molecule shown;
c peptide bond labelled between C of C=O and N of N-H;
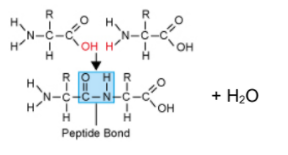
b.
a digested by peptidases/proteases;
b pepsin/pepsinogen/endopeptidase secreted by stomach (lining)/digests proteins in stomach;
c pancreas secretes/pancreatic juice contains endopeptidase/trypsin/peptidase;
d endopeptidase digest proteins/polypeptides to shorter chains of amino acids/shorter peptides;
e amino acids absorbed by active uptake/transport;
f in small intestine/ileum;
g villi increase the surface area for absorption;
h absorbed into bloodstream/into capillaries;
c
a mRNA is translated;
b mRNA binds with ribosome/with small subunit of ribosome;
c tRNA-activating enzymes/aminoacyl tRNA synthetases attach specific amino acid to tRNA;
d anticodon of 3 bases/nucleotides on tRNA;
e start codon/AUG on mRNA;
f tRNA carrying first amino acid/methionine binds to P/peptidyl site (when large subunit binds);
g anticodon (on tRNA) binds to codon (on mRNA);
h complementary base pairing (between codon and anticodon);
i tRNA for next codon binds to A site/amino acyl site;
j peptide bond forms between amino acids (on tRNAs) at P and A sites;
k ribosome moves along mRNA to next codon/by three bases/in 5’ to 3’direction;
ltRNA released from E/exit site;
m process/cycle repeats to elongate the polypeptide/until stop codon is reached;
n release of polypeptide and mRNA/disassembly of ribosome complex at stop codon;
