IB DP Biology Classification and cladistics Study Notes
IB DP Biology Classification and cladistics Study Notes at IITian Academy focus on specific topic and type of questions asked in actual exam. Study Notes focus on IB Biology syllabus with guiding questions of
- What tools are used to classify organisms into taxonomic groups?
- How do cladistic methods differ from traditional taxonomic methods?
- IB DP Biology 2025 SL- IB Style Practice Questions with Answer-Topic Wise-Paper 1
- IB DP Biology 2025 HL- IB Style Practice Questions with Answer-Topic Wise-Paper 1
- IB DP Biology 2025 SL- IB Style Practice Questions with Answer-Topic Wise-Paper 2
- IB DP Biology 2025 HL- IB Style Practice Questions with Answer-Topic Wise-Paper 2
A3.2.1 – Need for Classification of Organisms
🌍 Why Do We Need Classification?
The Earth is home to millions of species, and new ones are still being discovered. Without an organized system, studying or understanding this immense biodiversity would be chaotic.
🧭 Purpose of Classification
Classification is the systematic arrangement of organisms into groups based on shared characteristics.
✅ Main Reasons for Classification
| Purpose | Explanation |
|---|---|
| Simplifies study | Reduces complexity by organizing species into manageable categories |
| Identifies relationships | Reveals evolutionary and ecological connections between organisms |
| Predictive value | Allows scientists to predict characteristics shared by members of a group |
| Aids communication | Provides a universal language among scientists globally |
| Supports research | Organizes biological data, enabling comparison and discovery |
| Informs conservation | Helps prioritize protection of endangered species and ecosystems |
📌 Classification is essential for making sense of the vast diversity of life on Earth.
It groups organisms by similar traits, making biological study, communication, and conservation easier and more effective.
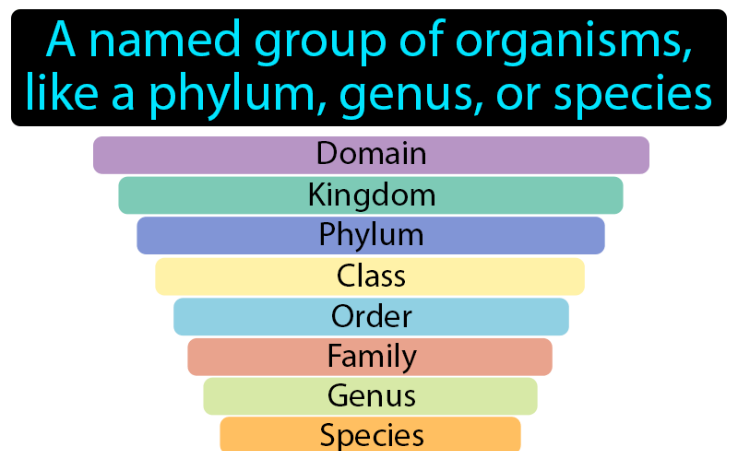
A3.2.2 – Difficulties Classifying Organisms into the Traditional Hierarchy of Taxa
🧬 What Is the Traditional Hierarchy of Taxa?
Biologists historically used a fixed system of classification based on ranked groups, known as taxa. The standard hierarchy includes:
Kingdom → Phylum → Class → Order → Family → Genus → Species
Each rank contains organisms with increasing similarity as you move down the list.
⚠️ Why Is This System Problematic?
| Issue | Explanation |
|---|---|
| Doesn’t always reflect evolutionary history | The hierarchy often fails to show true evolutionary relationships or timing of divergence |
| Taxonomic ranks are arbitrary | The boundaries between categories (e.g., what counts as a genus vs. family) are not fixed by nature |
| Gradual divergence complicates classification | Evolution is a gradual process, so species may not fit neatly into one rank or another |
| Disagreements among scientists | Taxonomists may disagree on where certain organisms fit, especially as new genetic data emerges |
🔍 Example of the Problem
Two species may appear very similar and be grouped into the same genus.
Over time, one evolves significantly. Should it still remain in the same genus?
There’s no objective rule – decisions become subjective.
💡 Alternative: Cladistics
| Traditional System | Cladistics |
|---|---|
| Uses fixed ranks (e.g., phylum, class) | Uses unranked clades based on common ancestry |
| Based on morphology | Based on genetic and evolutionary relationships |
| Ranks can be arbitrary | Clades are natural groups descended from a common ancestor |
🧠 Paradigm Shift in Biology
The move from traditional taxonomy to cladistics is a classic example of a paradigm shift where new scientific thinking replaces outdated models based on better evidence and logic.
📌 The traditional taxonomic hierarchy helps organize species but doesn’t always match evolutionary patterns.
Cladistics offers a more accurate alternative by focusing on common ancestry rather than rigid ranks.
A3.2.3 – Advantages of Classification Corresponding to Evolutionary Relationships
🌍 What is Evolutionary Classification?
- Evolutionary classification groups organisms based on shared ancestry, not just appearance.
- The ideal classification system reflects the evolutionary history of organisms.
- All members of a taxonomic group (like a genus or family) should have descended from a common ancestor.
🔑 Key Concepts
| Term | Definition |
|---|---|
| Clade | A group of organisms that includes an ancestor and all its descendants. |
| Synapomorphies | Shared features inherited from a common ancestor (e.g. feathers in birds). |
| Monophyletic group | A taxonomic group that includes only organisms that share a single common ancestor. |
🎯 Why Use Evolutionary Relationships for Classification?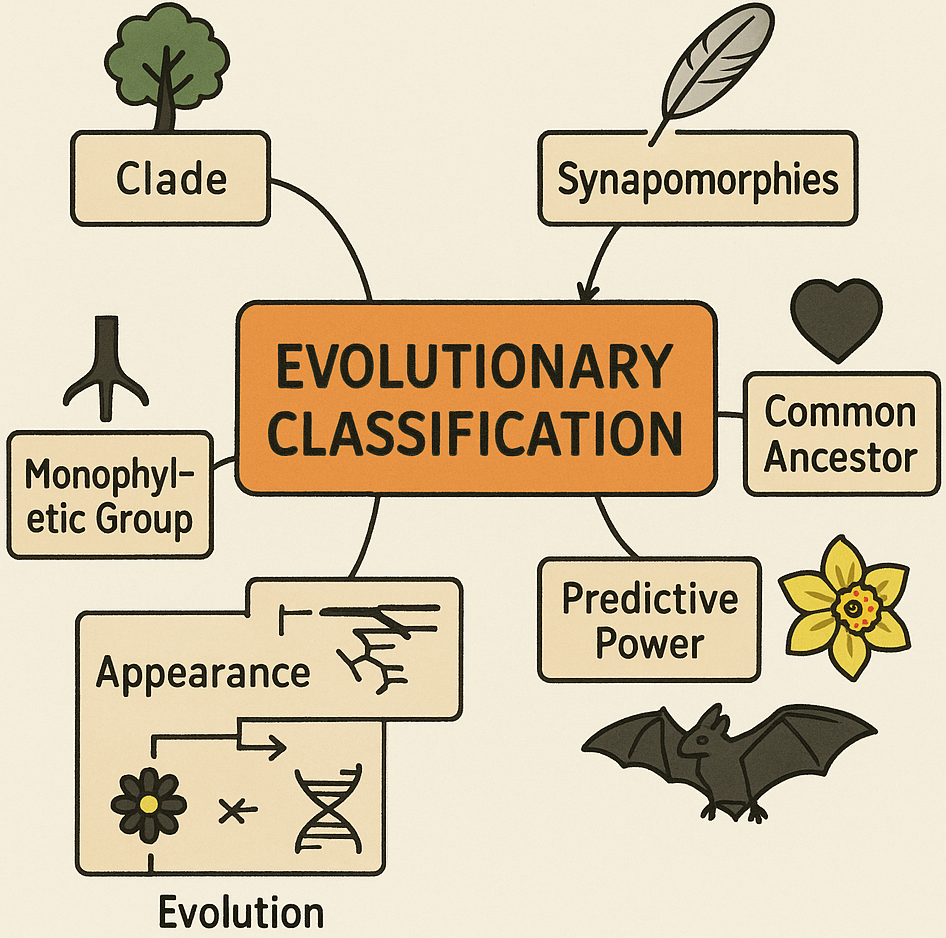
- Predictive Power: Traits can be predicted based on ancestry.
- Reflects true biological relationships.
- More accurate than appearance-based grouping.
- Supports research into genetics, evolution, ecology, and physiology.
🧬 Examples
🦇 Bats
Since bats are classified as mammals, it is predicted that:
- They have hair and mammary glands,
- A four-chambered heart,
- A placenta (and hence a navel),
- And other mammalian traits.
🌼 Daffodils (Narcissus species)
All species in the genus Narcissus descended from a common ancestor.
It is expected that all of them produce alkaloids, like galanthamine, used in Alzheimer’s treatment.
📌 Classifying organisms by their evolutionary relationships gives structure to biology.
It helps scientists understand how life diversified, what features to expect in related species, and how species are truly connected over time.
A3.2.4 – Clades as Groups of Organisms with Common Ancestry and Shared Characteristics
🧬 What is a Clade?
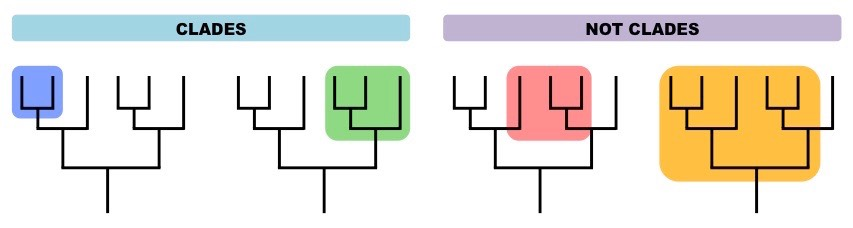
A clade is a group of organisms that:
- Evolved from a common ancestor
- Includes all descendants (living or extinct) of that ancestor
Note: Clades are also called monophyletic groups.
🌳 Cladograms: Evolutionary Trees
Cladograms are tree diagrams showing evolutionary relationships.
Each node (branch point) represents a speciation event (divergent evolution).
The branches show how species split and form new lineages over time.
📚 How Do We Identify Clades?
| Type of Evidence | Details |
|---|---|
| Genetic Evidence (Most reliable) | Compare base sequences of genes or amino acid sequences of proteins to identify similarities and shared ancestry. |
| Morphological Traits | Used when genetic data is unavailable (e.g. fossils). Involves comparing visible traits like limbs, leaves, etc. |
🧭 Clades Can Vary in Size
- Large Clades: e.g. Birds – over 10,000 species.
- Small Clades: e.g. Ginkgo biloba – the only living member of an ancient clade that evolved ~270 million years ago.
📦 Nested Clades
Every species belongs to multiple clades, with smaller ones nested inside larger groups.
Example: Gymnosperms (non-flowering seed plants)
- Araucaria araucana (monkey puzzle tree) and Podocarpus totara form a small clade.
- This clade is nested inside a larger one with Taxus baccata (yew).
- The entire group is part of a larger gymnosperm clade including Pinus radiata and even Ginkgo biloba.
🧠 Summary: Clades reflect true evolutionary relationships.
Genetic data gives the most objective evidence. Morphology is still useful, especially for extinct organisms.
Cladograms help visualize species’ ancestry and diversification.
A3.2.5 – Molecular Clock and Divergence of Clades
🧬 Key Concept: Gradual Genetic Differences
- Over time, mutations accumulate in the DNA and protein sequences of organisms.
- These sequence differences increase with time since two species diverged from a common ancestor.
Molecular Clock:
A method of estimating the time of divergence between species based on the rate of accumulation of genetic mutations.
🕒 How the Molecular Clock Works
- Assumes that mutations accumulate at a relatively constant rate over time.
- The more sequence differences, the longer ago two species shared a common ancestor.
- This provides a timeline of evolutionary history.
| Factor | Influence on Mutation Rate |
|---|---|
| Generation Time | Shorter generations → faster mutation accumulation |
| Population Size | Smaller populations can fix mutations more rapidly |
| Selective Pressure | High pressure can speed up or constrain evolution |
| Environmental Factors | Radiation, chemicals, or temperature can impact mutation rates |
🔍 Examples Using the Molecular Clock
| Species Compared | Estimated Divergence Time |
|---|---|
| Humans vs. Chimpanzees | ~4.5 million years ago |
| Chimpanzees vs. Bonobos | ~1 million years ago |
| All modern humans (mtDNA) | ~150,000 years ago |
💡 Limitations
- Mutation rates are not truly constant.
- Estimates are approximate and vary depending on:
- Calibration methods
- Type of DNA used (nuclear or mitochondrial)
- Evolutionary pressures in different lineages
Conclusion
The molecular clock is a powerful tool to estimate divergence times between species, but it must be interpreted cautiously due to variation in mutation rates.
A3.2.6 – Base Sequences or Amino Acid Sequences as the Basis for Constructing Cladograms
🧬 Why Use Base or Amino Acid Sequences?
All organisms share a universal genetic code, so differences in DNA or protein sequences can be used to:
- Compare species
- Determine evolutionary relationships
- Build cladograms (evolutionary trees)
🔁 Comparing Sequences to Infer Relatedness
| Type of Sequence | Example | Use in Cladistics |
|---|---|---|
| DNA base sequence | e.g., ATCG | Compare mutations or substitutions between species |
| Amino acid sequence | e.g., Met-Ala-Val | Compare proteins (e.g., hemoglobin) to infer relationships |
Fewer differences → more closely related (recent divergence)
More differences → more distantly related (earlier divergence)
🌳 Cladograms
Cladogram: A branching diagram showing shared ancestry.
Each branch point (node) represents a common ancestor.
Branch length may represent genetic distance or time since divergence.
⚙️ How Are Cladograms Constructed?
- Collect sequence data (DNA or amino acids) from species.
- Align sequences to compare similarities and differences.
- Use software tools to:
- Calculate all possible tree arrangements.
- Choose the most parsimonious tree.
📌 Parsimony Analysis
Parsimony = simplicity
The most likely cladogram is the one that:
- Explains observed differences with the smallest number of changes.
- Avoids assuming too many mutations.
Parsimony does not guarantee absolute truth, but suggests the most probable hypothesis.
🔍 Example (Simplified)
Suppose we compare a protein sequence in four species:
- Human: ATCGGA
- Chimpanzee: ATCGCA
- Gorilla: ATCCCA
- Orangutan: ATTCCA
→ The species with the least differences from humans (e.g., chimpanzees) are placed closer on the cladogram.
🧠 Nature of Science (NOS) Link
Scientific hypotheses may differ depending on:
- What data is used
- What criteria is applied
Different data sets or weighting of traits may lead to different cladograms.
Parsimony analysis is an example of how scientists balance complexity and evidence when interpreting evolutionary relationships.
A3.2.7 – Analysing Cladograms
🌳 What is a Cladogram?
A cladogram is a tree-like diagram used to show evolutionary relationships.
It reflects how organisms have diverged from common ancestors based on genetic or physical traits.
🔑 Key Terms in a Cladogram
| Term | Meaning |
|---|---|
| Root | The base of the cladogram – represents the most recent common ancestor of all organisms in the diagram. |
| Node | A branching point – shows where a single ancestral species split into two or more clades (groups). It represents a hypothetical common ancestor. |
| Terminal Branch | The end point of a branch – represents a current species or group of organisms. |
🧬 Understanding Relationships
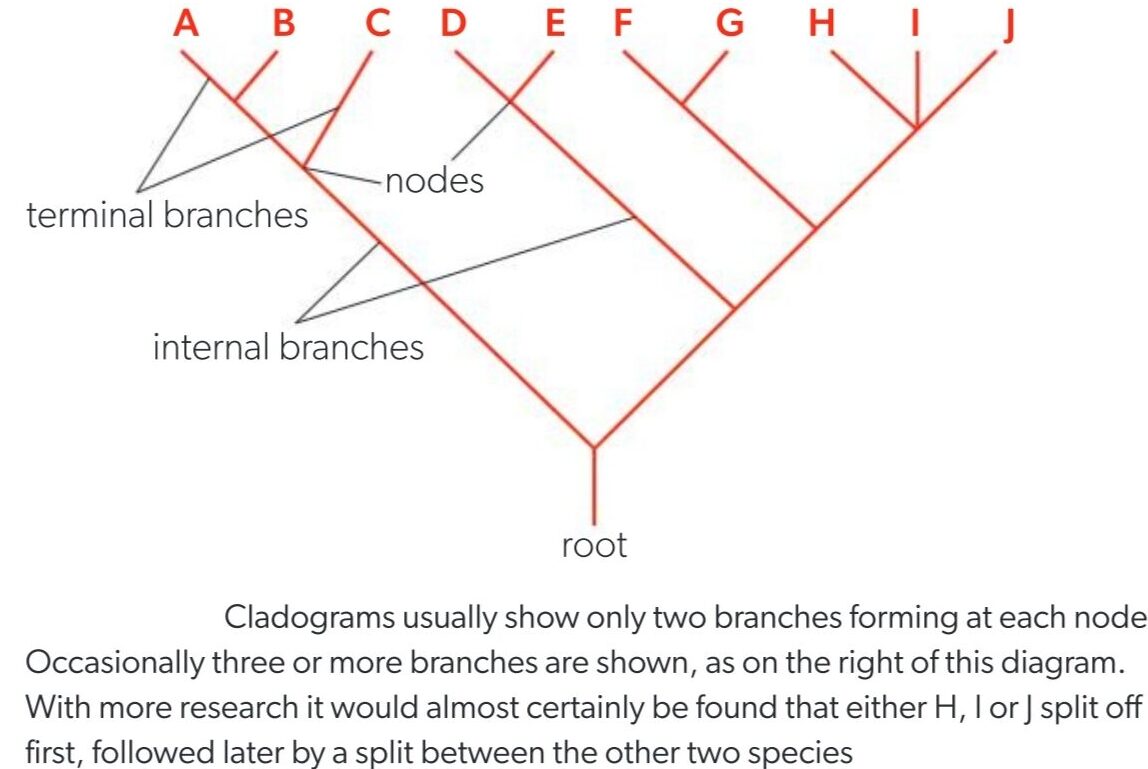
- Closer nodes = More closely related species.
- More nodes between groups = More distantly related.
- Species that branch from the same node are considered sister groups.
Cladograms may also include:
- Sequence differences (as numbers)
- Time estimates (length of branches, if scaled)
✅ How to Analyse a Cladogram
- Find the root – Identify the common ancestor of all species shown.
- Trace from node to tip – Follow the branches to see how species diverged.
- Compare clades – Species that share a more recent node are more closely related.
- Read terminal branches – These show the species being compared.
- Count nodes between species – More nodes = greater genetic divergence.
🔍 Example
If humans and chimpanzees are connected at one node, and that node connects to gorillas via another node:
- Humans and chimpanzees are closely related (recent divergence).
- Gorillas are less closely related (diverged earlier).
🧠 Tips: Always identify:
- Common ancestors (at nodes)
- Clades (each branch)
- Relative relatedness (who is closest to whom)
Use evidence from the diagram – number of nodes, branch length, or genetic differences (if provided).
A3.2.8 – Using Cladistics to Investigate Classification vs. Evolutionary Relationships
🧬 Cladistics and Evolution
Cladistics: A method of classification based on common ancestry and shared derived traits (synapomorphies).
It helps scientists determine whether the traditional classification of organisms reflects their true evolutionary history.
🧪 How Cladistics Tests Traditional Classification
| Aspect | Explanation |
|---|---|
| Traditional classification | Based mainly on morphology (observable traits). |
| Cladistic analysis | Based on genetic data and evolutionary lineage. |
| Result | Reveals if morphological similarities are due to common ancestry or convergent evolution. |
🔍 Example: Figwort Family (Scrophulariaceae)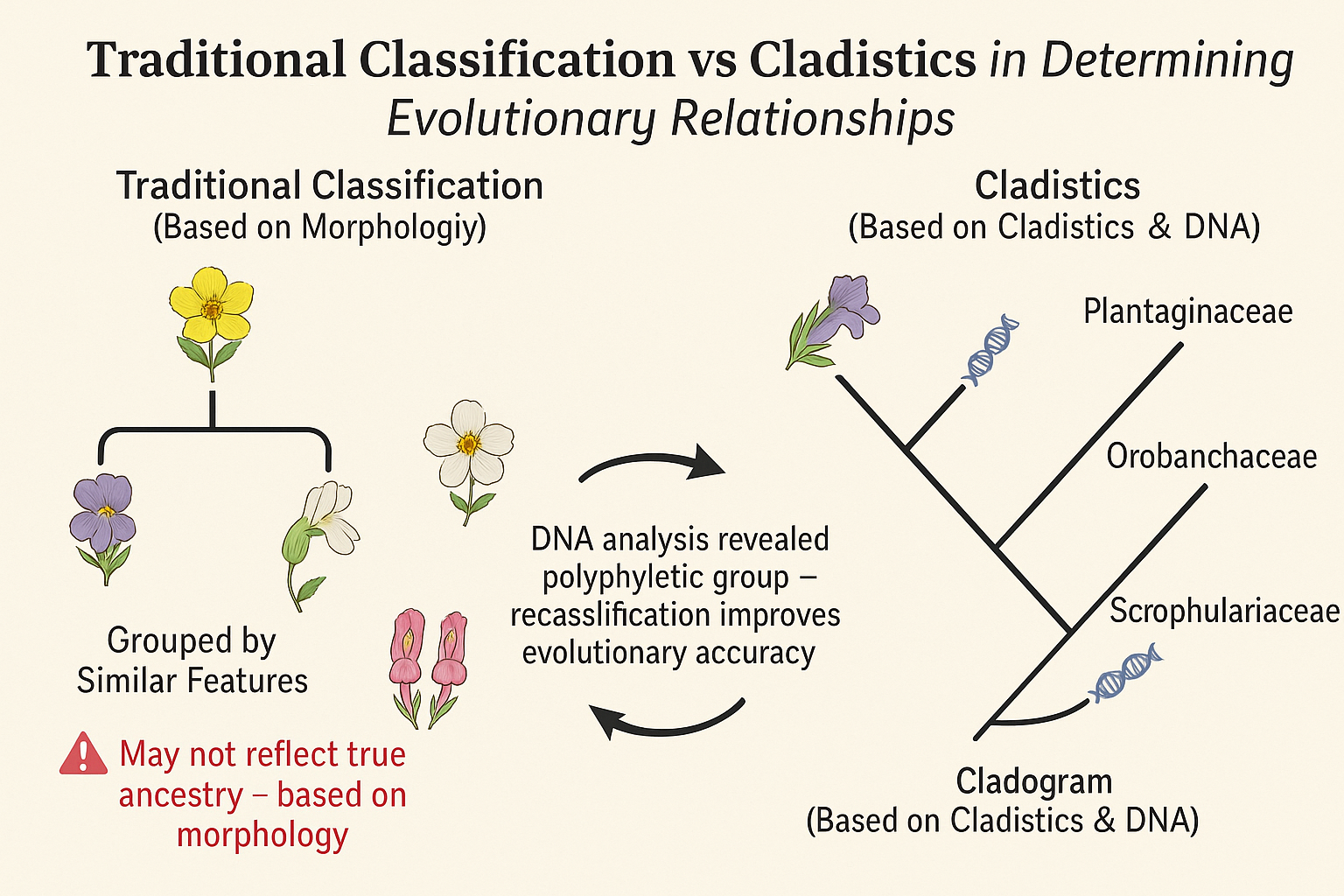
Originally, the figwort family included many species grouped based on similar flower structures.
Cladistic analysis (using DNA) revealed that these species did not all share a recent common ancestor.
Conclusion: The group was polyphyletic (made up of unrelated species).
Outcome: The figwort family was reclassified – some species were moved to Plantaginaceae and Orobanchaceae.
Note: You don’t need to memorize the figwort case, just understand that such reclassifications happen when cladistics exposes inaccurate groupings.
🔁 Why Reclassification Happens
Traditional taxonomy may rely on analogous structures (e.g., wings in bats and birds).
Cladistics highlights that these features evolved independently, not from a shared ancestor.
Therefore, classification based purely on looks can be misleading.
🧠 Nature of Science (NOS) Link
Scientific theories can be falsified when new evidence (e.g., genetic data) contradicts previous ideas.
The reclassification of plant families shows that:
- Scientific knowledge evolves with new tools and evidence.
- Classification systems are hypotheses open to revision.
✅ Key Takeaways:
- Cladistics tests whether species in a group truly share a common ancestor.
- It uses DNA sequences to provide more accurate evolutionary trees.
- Misclassified organisms are reassigned to reflect their true evolutionary relationships.
- This improves the accuracy and usefulness of biological classification systems.
A3.2.9 – Classification of All Organisms into Three Domains Using rRNA Base Sequences
🔹 Traditional View (Before 1977)
Organisms were grouped into:
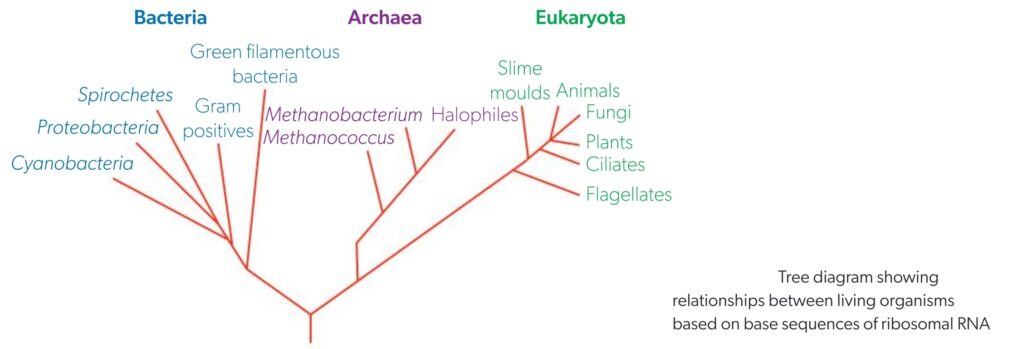
- Prokaryotes → No nucleus
- Eukaryotes → With nucleus
This classification was oversimplified and didn’t reflect actual evolutionary relationships.
🔹 Revolutionary Change (1977 – Carl Woese)
rRNA sequencing revealed deep genetic differences among prokaryotes.
Result: Prokaryotes split into two distinct groups:
- Eubacteria
- Archaea
🔹 Three-Domain System
Now all life is classified into three domains (a level above kingdoms):
| Domain | Key Features | Examples |
|---|---|---|
| Bacteria | True bacteria, prokaryotic, diverse metabolism | E. coli, Streptococcus |
| Archaea | Extremophiles, unique genes & membranes | Methanogens, Halophiles |
| Eukaryota | Cells with nuclei and organelles | Plants, Animals, Fungi, Protists |
🔹 Why Use rRNA?
- Present in all living cells
- Slow to change → Good for comparing distant organisms
- Provides molecular evidence of ancestry and divergence
🔹 Nature of Science (NOS) Link
This classification shift was a paradigm shift in biology — based on molecular data rather than visible traits.
It demonstrates how new evidence can redefine scientific understanding and reshape classification systems.
