Question
(a) Organisms can be classified by their features and by studying the sequence of bases in their DNA.
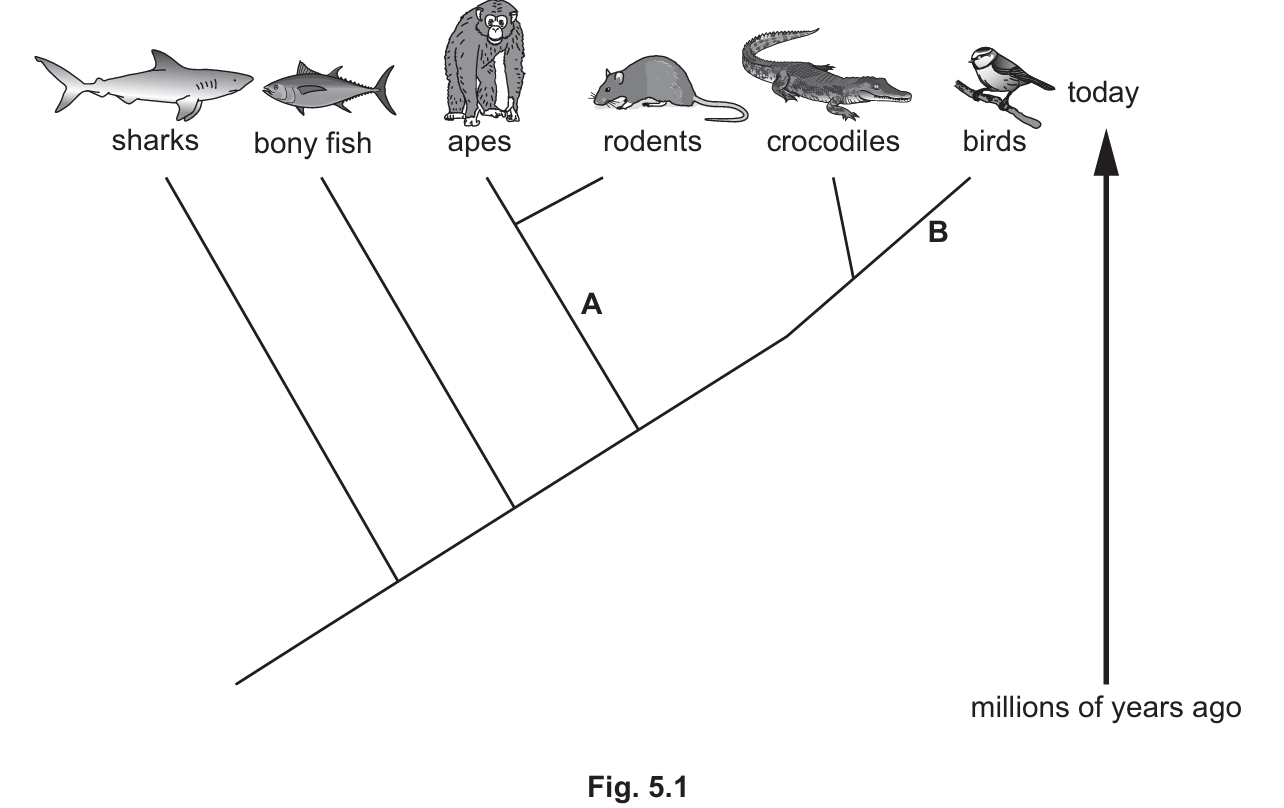
Fig. 5.1 is a diagram showing the evolutionary relationships between some different groups of organisms.
Each branch shows the point at which organisms developed new features that classify them as a new group.
The point where the branch starts also indicates a common ancestor shared by the new groups.
▶️ Answer/Explanation
(a)(i)
A: Hair / external ears (pinnae)
B: Wings / feathers / beaks
Explanation: Point A is the branch leading specifically to apes and rodents. Both of these groups belong to the class Mammalia, and the defining visible features of mammals are the presence of hair/fur and external ears. Point B is the branch leading to birds. The defining visible features for this class are feathers and wings (modified forelimbs).
(a)(ii)
Apes and rodents
Explanation: On an evolutionary tree (cladogram), the most recent common ancestor is represented by the node (branching point) closest to the species in question. Apes and rodents originate from the same specific branch point (labeled A), indicating they split from each other more recently than they split from reptiles, fish, or sharks.
(a)(iii)
Most similar: Birds
Least similar: Sharks
Explanation: Classification reflects evolutionary relationships. Organisms that share a more recent common ancestor have more similar DNA base sequences.
- Birds share the closest node (node B) with crocodiles, indicating they are the closest relatives on this diagram.
- Sharks branched off the earliest (at the far left of the diagram), meaning they are the most distantly related to crocodiles and will have the most accumulated differences in their DNA sequences.
(b)
A DNA molecule consists of two strands (or chains) coiled together to form a double helix. Each strand contains chemicals called bases. There are four bases: Adenine (A), Thymine (T), Cytosine (C), and Guanine (G). These bases pair up in a specific way: A pairs with T and C pairs with G. The strands are held together by bonds between these base pairs.
Explanation: This question requires recalling the fundamental structure of DNA as outlined in syllabus section 4.1. Key points are the double-stranded nature, the specific “twisted ladder” shape (double helix), and the complementary base pairing rules.
(c)
The sequence of bases in DNA (or the mRNA copy) determines the sequence of amino acids. These amino acids are joined together to form proteins (a process called protein synthesis). The specific sequence of amino acids determines the shape of the protein, and the shape determines its function (e.g., the active site of an enzyme). Therefore, by coding for specific proteins, DNA controls cell function.
Explanation: This connects the genetic code (genotype) to the physical traits and functions (phenotype). The “Central Dogma” of biology flows from DNA $\rightarrow$ mRNA $\rightarrow$ Amino Acid Sequence $\rightarrow$ Protein $\rightarrow$ Cell Function.
