NEET Biology - Unit 7- Molecular basis of Inheritanc- Study Notes - New Syllabus
NEET Biology – Unit 7- Molecular basis of Inheritanc- Study Notes – New Syllabus
Key Concepts:
- Molecular basis of Inheritance: Search for genetic material and DNA as genetic material; Structure of DNA and RNA; DNA packaging; DNA replication; Central dogma; Transcription, genetic code, translation; Gene expression and regulation- Lac Operon; Genome and human genome project; DNA finger printing, protein biosynthesis.
Molecular Basis of Inheritance
🌿 Introduction
Inheritance in living organisms is controlled by genetic material, which stores and transmits information from one generation to another. The search for genetic material began in early 20th century and led to the discovery of DNA as the carrier of hereditary information.
🔎 1. Search for Genetic Material
1.1 Griffith’s Experiment (1928) – Transforming Principle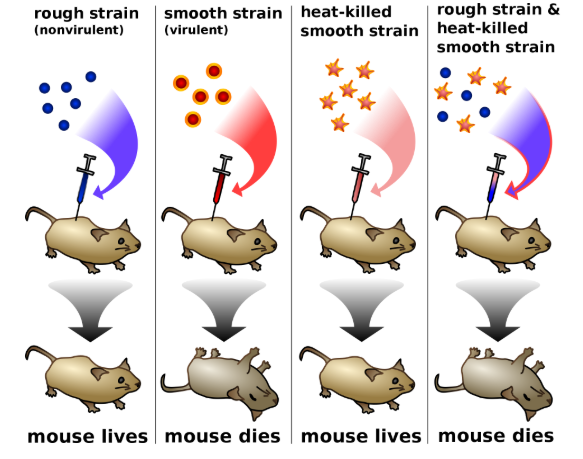
- Studied Streptococcus pneumoniae:
- S strain (Smooth) → polysaccharide coat, virulent, kills mice
- R strain (Rough) → lacks coat, non-virulent, mice survive
- Experiment Results:
- Studied Streptococcus pneumoniae:
| Experiment | Result |
|---|---|
| S strain injected into mice | Mice die |
| R strain injected | Mice survive |
| Heat-killed S strain injected | Mice survive |
| Heat-killed S + live R strain | Mice die; living S recovered |
- Conclusion: Something from heat-killed S strain transformed R strain into virulent bacteria → “Transforming Principle”
1.2 Avery, MacLeod & McCarty (1944) – DNA as Transforming Principle
- Isolated DNA, RNA, proteins from S strain
- Only DNA converted R → S strain; proteins and RNA had no effect
- Conclusion: DNA is the biochemical substance responsible for heredity
1.3 Hershey-Chase Experiment (1952) – Viral DNA is Genetic Material
- Bacteriophages infecting E. coli
- Labelled viral components: DNA (P³²), Protein coat (S³⁵)
 Observations:
Observations:- Only bacteria infected with radioactive DNA became radioactive → new viruses produced
- Bacteria infected with radioactive protein did not become radioactive
- Conclusion: DNA, not protein, carries genetic instructions
1.4 RNA World Hypothesis
- RNA may have been first genetic material
- Functioned as catalyst and information carrier
- RNA reactive → DNA evolved for stability and repair
- DNA → double-stranded → stable, stores information safely
🧬 2. DNA – The Genetic Material
2.1 Historical Milestones
- 1869: Miescher → isolated nuclein (DNA)
- 1953: Watson & Crick → double helix structure
2.2 Definition
“Deoxyribonucleic acid (DNA) is a molecule that carries genetic instructions used in growth, development, functioning, and reproduction of all living organisms and many viruses.”
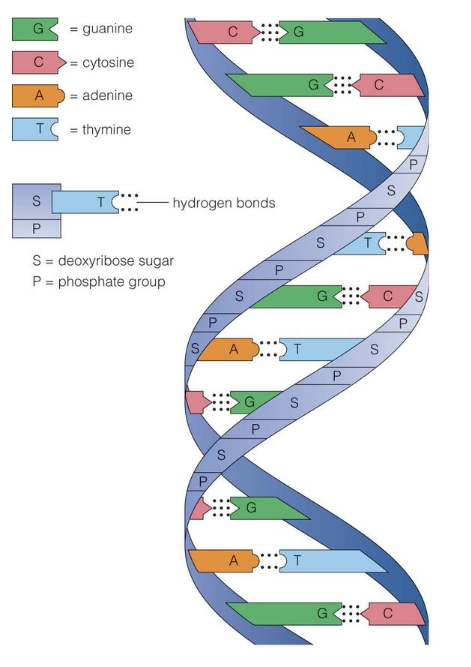
2.3 Structure of DNA
- Composition: polymer of deoxyribonucleotides
- Nucleotide components: Nitrogenous base (A, T, G, C), Sugar (deoxyribose), Phosphate group
- Polynucleotide Chain: sugar-phosphate backbone, bases inside
- Two antiparallel strands → double helix
- Base Pairing: A-T (2 H-bonds), G-C (3 H-bonds); purine ↔ pyrimidine

- Double Helix Features: right-handed, 10 bp/turn, major & minor grooves, genetic info on sense/template strand
🏷 3. Packaging of DNA
- Prokaryotes: DNA held in nucleoid, organized into loops
- Eukaryotes: DNA packaged into chromatin
- Histones: H2A, H2B, H3, H4 → octamer → nucleosome (~200 bp, beads-on-string)
- Non-histone proteins → higher-order packaging
- Euchromatin: loosely packed, active; Heterochromatin: tightly packed, inactive
🧬 4. DNA Replication
- Definition: DNA replication is the process by which DNA makes a copy of itself during cell division
- Steps of Replication:
- Unwinding: Helicase → replication fork
- Primer Formation: Primase synthesizes RNA primers
- Strand Synthesis: Leading strand continuous, lagging strand → Okazaki fragments
- Primer Removal & Gap Filling: DNA polymerase fills gaps
- Joining Fragments: DNA ligase seals fragments
- Proofreading: DNA polymerase ensures accuracy
- Semiconservative Replication: Each DNA = one old + one new strand
📘 5. Summary Table
| Feature | Details |
|---|---|
| DNA | Double-stranded, antiparallel, double helix |
| Bases | A-T (2 H-bonds), G-C (3 H-bonds) |
| Backbone | Sugar-phosphate, covalent bonds |
| Packaging | Nucleosome → beads-on-string (eukaryotes), loops in prokaryotes |
| Genetic material evidence | Griffith → transforming principle, Avery → DNA, Hershey-Chase → DNA confirmed |
| Replication | Semiconservative, leading & lagging strands, Okazaki fragments, DNA ligase seals, proofreading ensures accuracy |
🧾 Quick Recap
DNA is the genetic material → double-stranded, A-T & G-C pairing
Discovered by Miescher, structure by Watson & Crick
Packaging: nucleosome → histone octamer → beads-on-string
Evidence: Griffith (1928), Avery-MacLeod-McCarty (1944), Hershey-Chase (1952)
Replication: semiconservative → helicase unzips, primers added, DNA polymerase synthesizes, ligase seals
Molecular Basis of Inheritance: DNA and RNA Structure & DNA Packaging
🌿 1. DNA (Deoxyribonucleic Acid)
1.1 Definition
“Deoxyribonucleic acid (DNA) is a molecule that carries the genetic instructions required for growth, development, functioning, and reproduction of all living organisms and many viruses.”
DNA is a polymer of nucleotides. Length depends on the number of nucleotides. Most DNA exists as double-stranded helix.
1.2 Components of DNA
- Nitrogenous Base: Purines: Adenine (A), Guanine (G); Pyrimidines: Thymine (T), Cytosine (C)
- Sugar: Deoxyribose (pentose sugar)
- Phosphate Group: Forms covalent phosphodiester bonds with sugar of next nucleotide
- Polynucleotide Chain: Sugar-phosphate backbone outside; bases point inwards → store genetic information
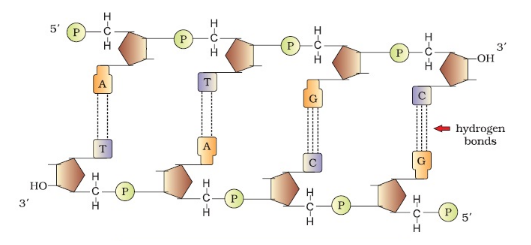
1.3 Structure of DNA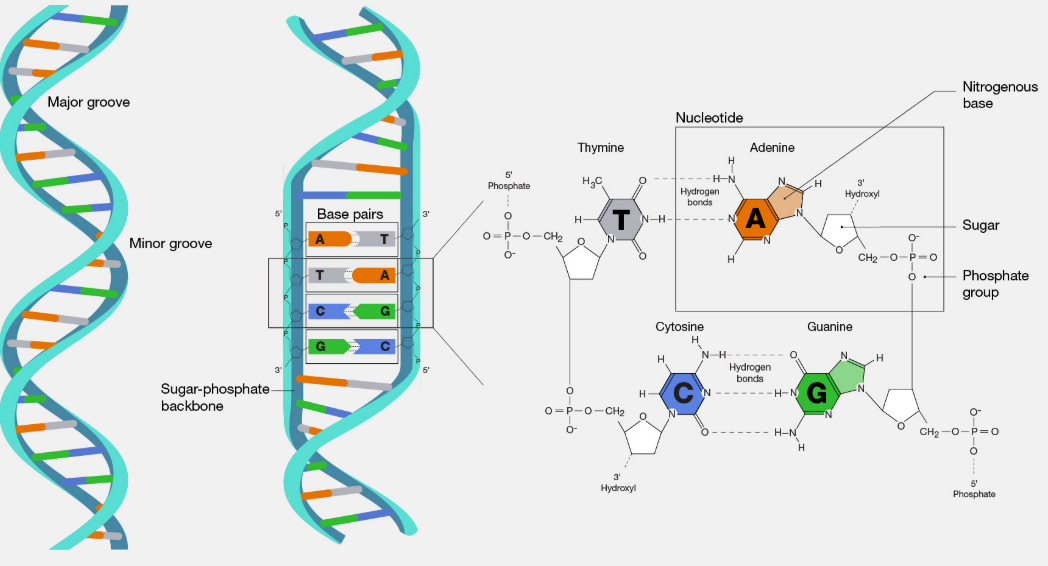
- Double Helix Features: two polynucleotide chains, antiparallel (5’→3’ & 3’→5’)
- Base Pairing (Chargaff’s Rule): A ↔ T (2 H-bonds), G ↔ C (3 H-bonds)
- Helix properties: Right-handed, pitch = 3.4 nm, 10 base pairs/turn, distance between base pairs = 0.34 nm
- Stabilization: base stacking + H-bonds; major & minor grooves → protein binding & gene regulation
- Sense strand/template strand: carries genetic code
- Summary Diagram (descriptive): Two polynucleotide chains coiled → sugar-phosphate backbone outside, bases inside → A-T, G-C complementary pairing → antiparallel orientation → double helix
1.4 Salient Features of DNA
- Stores biological information; resistant to cleavage
- Two strands store same info → allows accurate replication
- Purine pairs with pyrimidine
- Helix stabilized by H-bonds and base stacking
- Major and minor grooves → regulatory protein binding
🌿 2. RNA (Ribonucleic Acid)
2.1 Definition
RNA is a nucleic acid that helps in the expression of genetic information stored in DNA.
2.2 Components of RNA
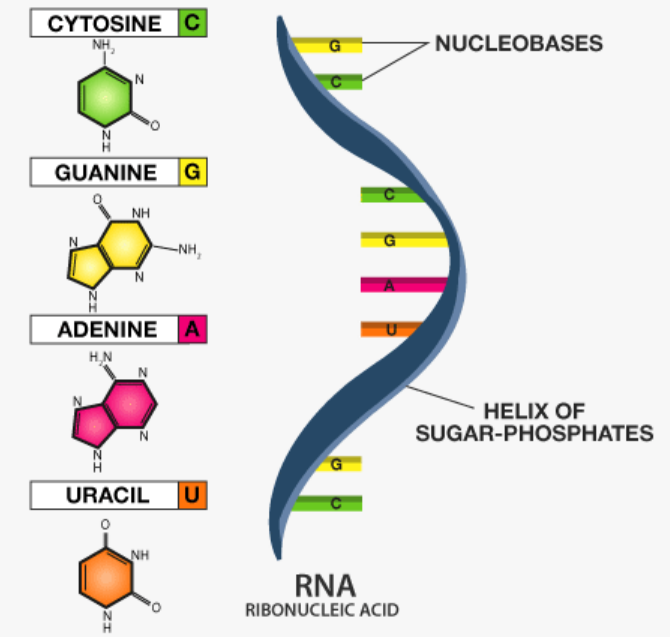
- Nucleotides: Nitrogenous bases → A, U, G, C (Uracil replaces Thymine)
- Sugar: Ribose
- Phosphate group
2.3 Structure of RNA
- Single-stranded molecule
- Forms secondary structures: hairpins, loops (intramolecular base pairing)
- Types:
- mRNA: carries info from DNA → ribosome
- tRNA: carries amino acids for protein synthesis
- rRNA: structural & functional component of ribosome
- Less stable than DNA due to reactive OH group on ribose
Key Differences: DNA vs RNA
| Feature | DNA | RNA |
|---|---|---|
| Strands | Double-stranded | Single-stranded |
| Sugar | Deoxyribose | Ribose |
| Bases | A, T, G, C | A, U, G, C |
| Function | Genetic info storage | Gene expression & protein synthesis |
| Stability | Stable | Less stable, reactive |
🌿 3. DNA Packaging
3.1 Packaging in Prokaryotes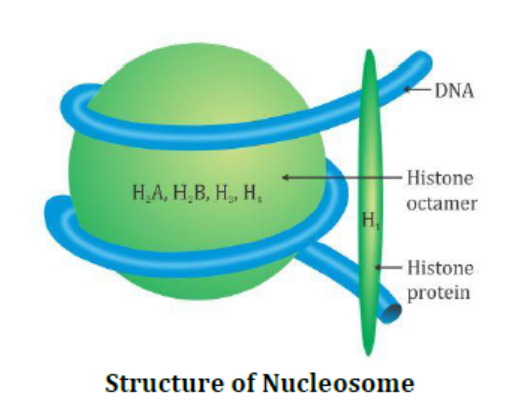
- Circular DNA located in nucleoid
- Loops stabilized by proteins
- No true nucleus
3.2 Packaging in Eukaryotes
- Linear DNA packaged into chromatin
- Histone proteins: H2A, H2B, H3, H4 (2 each) → octamer
- DNA wraps around octamer → nucleosome (~200 bp) → repeating units → “beads-on-string” appearance
3.3 Higher-Order Packaging
- Nucleosomes coil → 30 nm fiber → loops/domains → chromosome
- Non-histone proteins stabilize higher-order chromatin
3.4 Chromatin Types
| Chromatin | Features |
|---|---|
| Euchromatin | Loosely packed, transcriptionally active |
| Heterochromatin | Densely packed, transcriptionally inactive, stains dark |
Key Point: DNA packaging ensures efficient storage in nucleus and regulates gene expression.
Difference between DNA and RNA
| DNA | RNA |
|---|---|
| DNA occurs inside nucleus and in some organelles like mitochondria and chloroplast. | Very little RNA is present in nucleus; majority is found in cytoplasm. |
| It is double stranded (exception: some viruses). | It is single stranded (exception: Reovirus). |
| Composed of millions of nucleotides. | Depending on type, RNA has 70 to 12,000 nucleotides. |
| Contains 2-deoxyribose sugar. | Contains ribose sugar. |
| Purine and pyrimidine bases are equal. | No fixed ratio between purine and pyrimidine bases. |
| Bases: Adenine (A), Guanine (G), Thymine (T), Cytosine (C). | Bases: Adenine (A), Guanine (G), Uracil (U), Cytosine (C). |
| Hydrogen bonds form between A-T and C-G on opposite strands. | Base pairing occurs only in folded/coiled regions. |
| Forms a regular double helix. | May fold to form secondary or pseudo helices. |
| Replicates to form new DNA molecules. | Cannot replicate independently. |
| Occurs as chromosomes or chromatin. | Found in ribosomes or associated with ribosomes. |
| Transfers genetic information across generations. | Directs protein synthesis. |
| Quantity remains fixed. | Quantity varies with cell activity. |
| Two types: Intranuclear and extranuclear. | Three types: mRNA, tRNA, rRNA. |
| Long-lived molecule. | Short-lived molecule. |
🧾 Quick Recap
DNA: double-stranded, A-T & G-C pairing, antiparallel, double helix, sense strand → genetic info
RNA: single-stranded, U replaces T, types → mRNA, tRNA, rRNA, gene expression
DNA Packaging: Prokaryotes → nucleoid, loops with proteins; Eukaryotes → nucleosomes → chromatin → chromosomes
Euchromatin → transcriptionally active, Heterochromatin → inactive
DNA Replication
Definition
DNA replication is the process by which DNA makes an exact copy of itself during cell division, ensuring genetic information is passed to daughter cells.
Key Features of DNA Replication
- Semiconservative: Each new DNA molecule has one old strand + one newly synthesized strand
- Occurs in S-phase of cell cycle
- Template-based: Existing DNA strands act as templates for new strands
- Bidirectional: Replication occurs in both directions from origin of replication
Steps of DNA Replication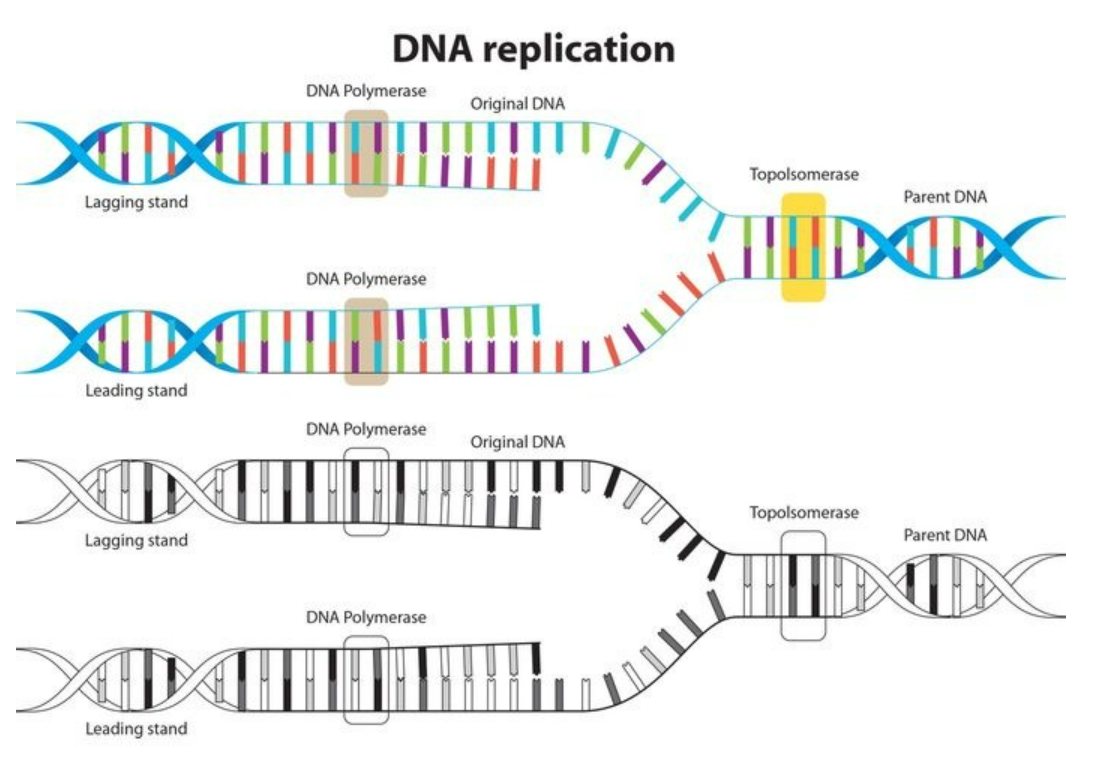
1. Initiation – Unwinding of DNA
- Enzyme: Helicase → breaks hydrogen bonds → forms replication fork (Y-shaped)
- Single-strand binding proteins (SSBPs) stabilize unwound DNA
- Diagram in words: Double helix → helicase opens it → two single-stranded templates ready
2 Primer Synthesis
- DNA polymerase cannot start new strand → requires primer
- Enzyme: Primase → synthesizes short RNA primer complementary to template strand
3 Elongation – Synthesis of DNA Strands
- Leading Strand (3’→5’ template): continuous synthesis 5’→3’ toward fork
- Lagging Strand (5’→3’ template): discontinuous → Okazaki fragments, each starts with RNA primer
4 Primer Removal & Gap Filling
- RNA primers removed by DNA polymerase exonuclease activity
- Gaps filled with DNA nucleotides complementary to template
5 Joining Fragments
- Enzyme: DNA ligase → seals nicks between Okazaki fragments → forms continuous strand
6 Proofreading and Error Correction
- DNA polymerase has 3’→5’ exonuclease activity → removes mispaired nucleotides
- Mutation rate is extremely low → ensures high-fidelity replication
4. Enzymes Involved in DNA Replication
| Enzyme | Function |
|---|---|
| Helicase | Unwinds double helix |
| Primase | Synthesizes RNA primer |
| DNA Polymerase III | Adds nucleotides to growing DNA strand |
| DNA Polymerase I | Removes RNA primer, fills gaps with DNA |
| DNA Ligase | Joins Okazaki fragments |
| Topoisomerase / Gyrase | Relieves supercoiling ahead of fork |
| SSBPs | Stabilize single-stranded DNA |
5. Replication Models
- Semiconservative (Watson & Crick) → one old + one new strand
- Conservative → old DNA intact, new DNA entirely new (wrong)
- Dispersive → old and new DNA mixed (wrong)
- Meselson-Stahl Experiment (1958) → confirmed semiconservative replication using N¹⁵ isotope in E. coli
6. Features of DNA Replication
- Bidirectional from origin
- Semi-discontinuous: continuous leading, discontinuous lagging
- High fidelity due to proofreading
- Requires energy from dNTP hydrolysis
- Universal in all organisms
7. Summary of Replication Process
- Origin Recognition → helicase unwinds DNA → replication fork
- Primer Formation → primase adds RNA primer
- Elongation → DNA polymerase adds nucleotides
- Leading strand → continuous
- Lagging strand → Okazaki fragments
- Primer Removal & Gap Filling → DNA polymerase I
- Joining Fragments → DNA ligase
- Proofreading → DNA polymerase exonuclease activity ensures accuracy
🧾 Quick Recap
DNA replication is semiconservative → each daughter = 1 old + 1 new strand
Occurs at replication fork; bidirectional
Leading strand → continuous, Lagging strand → Okazaki fragments
Enzymes: Helicase, Primase, DNA polymerase, Ligase, Topoisomerase, SSBPs
Proofreading → ensures high fidelity, mistakes removed
Confirmed by Meselson-Stahl experiment
Central Dogma of Molecular Biology
Definition
- The Central Dogma describes the flow of genetic information in a cell: DNA → RNA → Protein
- DNA stores genetic instructions
- RNA acts as messenger
- Proteins perform cellular functions
- Proposed by Francis Crick (1958)
Steps in Central Dogma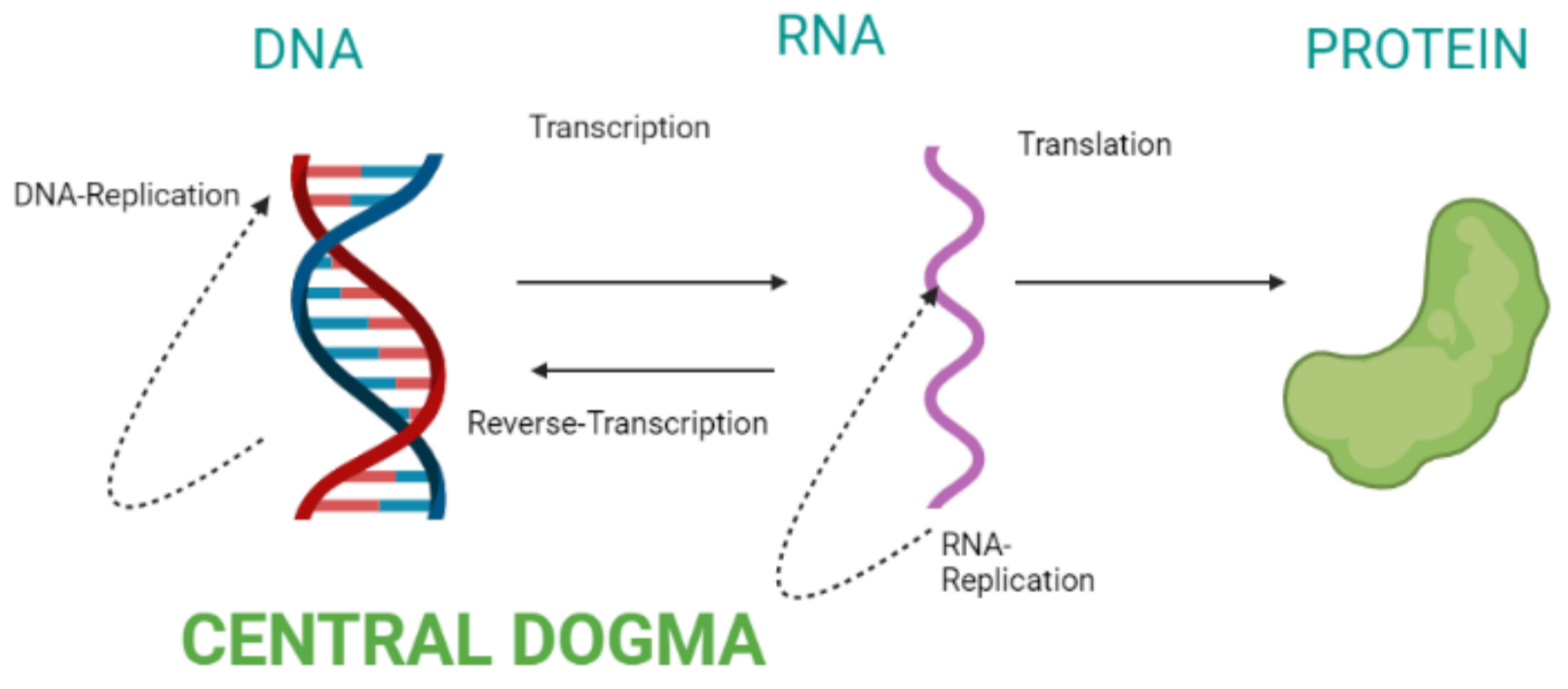
1. DNA Replication
- DNA copies itself → ensures genetic continuity
- Occurs before cell division (S-phase)
- Semiconservative: one old + one new strand
- Enzymes: Helicase, Primase, DNA polymerase, Ligase, Topoisomerase
- Flow: DNA → DNA
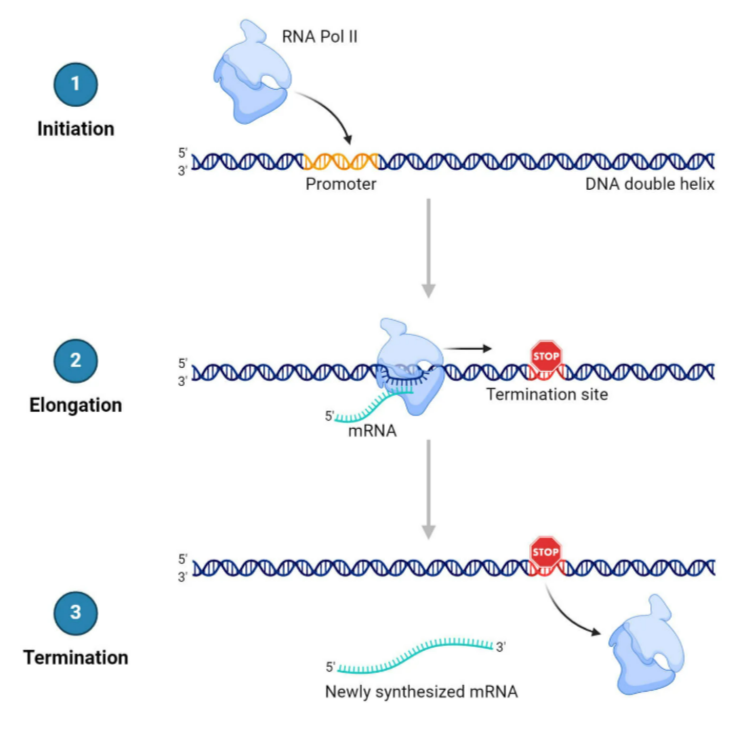
2. Transcription
- Process of synthesizing RNA from DNA template
- Occurs in nucleus (eukaryotes) or cytoplasm (prokaryotes)
- Steps:
- Initiation: RNA polymerase binds promoter
- Elongation: RNA polymerase synthesizes mRNA (5’→3’)
- Termination: RNA polymerase detaches at terminator
- Result: Pre-mRNA → mature mRNA (splicing, capping, poly-A tail)
- Flow: DNA → mRNA
3. Translation
- Process of synthesizing protein from mRNA
- Occurs at ribosomes in cytoplasm
- Requires tRNA, rRNA, mRNA
- Steps:
- Initiation: Ribosome binds start codon (AUG) on mRNA
- Elongation: tRNA brings amino acids → peptide bonds form
- Termination: Ribosome reaches stop codon (UAA, UAG, UGA) → polypeptide released
- Flow: mRNA → Protein
Types of RNA Involved
| RNA Type | Function |
|---|---|
| mRNA | Carries genetic info from DNA to ribosome |
| tRNA | Transfers amino acids to ribosome during protein synthesis |
| rRNA | Forms core structure of ribosome |
🧬 Genetic Code
- Set of rules to translate nucleotide sequence → amino acid sequence
- Triplet code: 3 nucleotides = 1 codon → 1 amino acid
- Features:
- Universal: almost all organisms
- Degenerate: multiple codons code same amino acid
- Start codon: AUG → Methionine
- Stop codons: UAA, UAG, UGA → termination
Overall Flow of Information
Diagram in words: DNA → transcription → mRNA → translation → Protein
Proteins carry out cellular structure & functions, completing the expression of genetic information
🧾 Quick Recap
Central Dogma: DNA → RNA → Protein
Replication: DNA → DNA (semiconservative, S-phase)
Transcription: DNA → mRNA (RNA polymerase, promoter, terminator, splicing)
Translation: mRNA → Protein (ribosome, tRNA, start/stop codon, peptide bond formation)
RNA types: mRNA, tRNA, rRNA
Genetic code: Triplet, universal, degenerate, start AUG, stop UAA/UAG/UGA
Transcription, Genetic Code, Translation
Definition
Transcription is the process of synthesizing RNA from a DNA template.
- Transfers genetic information from DNA → RNA
- Essential for protein synthesis
- Catalyzed by RNA polymerase
🌿 Site of Transcription
| Organism | Site |
|---|---|
| Prokaryotes | Cytoplasm (no nucleus) |
| Eukaryotes | Nucleus → processed before leaving to cytoplasm |
Types of RNA Produced
| RNA Type | Function |
|---|---|
| mRNA | Carries genetic code from DNA to ribosome |
| tRNA | Transfers amino acids to ribosome |
| rRNA | Ribosome structure & catalysis |
| snRNA | Splicing of pre-mRNA (eukaryotes) |
| miRNA/siRNA | Regulate gene expression |
Steps of Transcription
Step 1: Initiation
- RNA polymerase binds to promoter region of DNA
- DNA unwinds → forms transcription bubble

- Transcription factors help polymerase binding in eukaryotes
- Determines which gene is transcribed
Step 2: Elongation
- RNA polymerase moves along template strand (3’→5’)
- Synthesizes RNA 5’→3’
- Base pairing rules:
- DNA A → RNA U
- DNA T → RNA A
- DNA G → RNA C
- DNA C → RNA G
Step 3: Termination
- RNA polymerase reaches terminator sequence
- RNA transcript released
- DNA rewinds to original double helix
Post-Transcriptional Modifications (Eukaryotes)
- 5’ Cap: Added to 5’ end → protects RNA & helps ribosome binding
- 3’ Poly-A Tail: Added to 3’ end → prevents degradation
- Splicing: Removes introns, joins exons → mature mRNA
Note: Prokaryotic mRNA does not require modification
Important Points
- Only one DNA strand (template strand) is used for RNA synthesis
- Other strand → coding strand (same sequence as RNA except T → U)
- Transcription is gene-specific
- Produces complementary RNA strand
🔗Genetic Code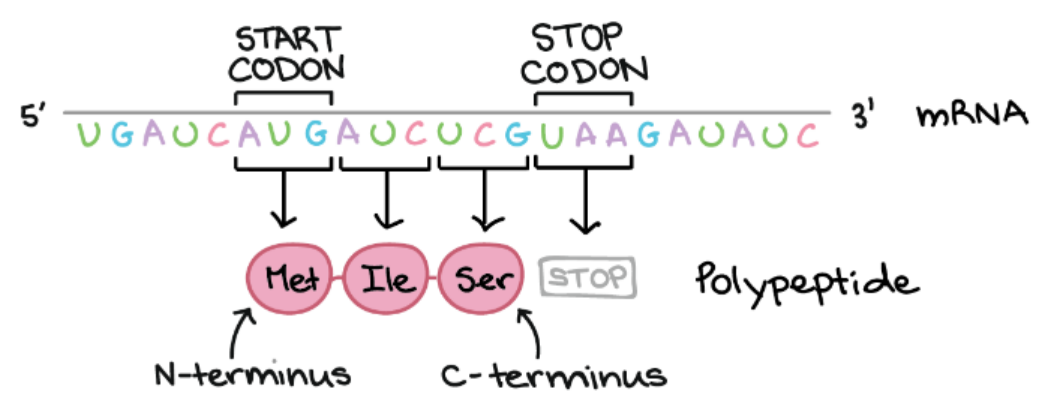
Definition
Genetic code is the set of rules by which the sequence of nucleotides in mRNA is translated into amino acids in a protein.
- DNA → RNA → Protein (Central Dogma)
- Codons in mRNA determine the order of amino acids
Features of Genetic Code
| Feature | Description |
|---|---|
| Triplet code | 3 nucleotides (codon) = 1 amino acid |
| Start codon | Signals start of translation: AUG → Methionine |
| Stop codon | Signals termination of translation: UAA, UAG, UGA |
| Degenerate / Redundant | Multiple codons code for same amino acid |
| Non-overlapping | Codons are read sequentially, no overlap |
| Universal | Same codons in almost all organisms |
| Comma-free | No pauses or punctuation between codons |
Types of Codons
| Codon Type | Function | Example |
|---|---|---|
| Start codon | Initiates translation | AUG → Methionine |
| Stop codon | Terminates translation | UAA, UAG, UGA |
| Sense codon | Codes for amino acid | GGC → Glycine |
| Nonsense codon | Does not code for amino acid | UAA, UAG, UGA |
Codon Examples
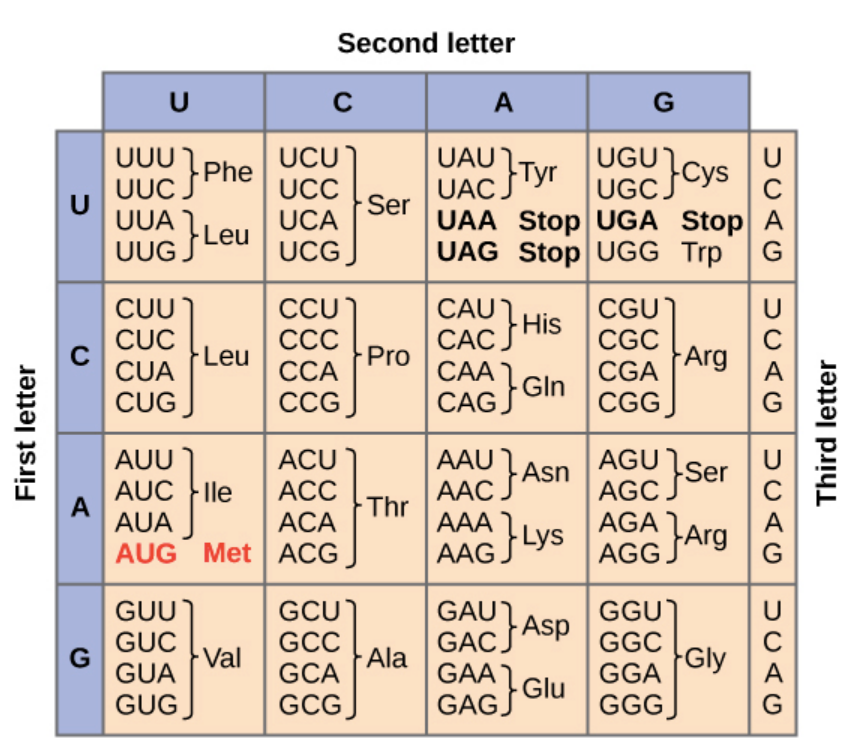
| Amino Acid | Codon(s) |
|---|---|
| Methionine (Start) | AUG |
| Phenylalanine | UUU, UUC |
| Glycine | GGU, GGC, GGA, GGG |
| Serine | UCU, UCC, UCA, UCG, AGU, AGC |
| Stop codon | UAA, UAG, UGA |
Example:
mRNA: 5’–AUG–GGC–UAC–3’ → Protein: Met–Gly–Tyr
Important Points
- Genetic code is universal, degenerate, and non-overlapping
- One codon → one amino acid
- Start codon AUG sets the reading frame
- Stop codons UAA, UAG, UGA do not code for amino acids
🧬Translation (Protein Synthesis)
Definition
Translation is the process of synthesizing a polypeptide (protein) from an mRNA template.
- Genetic information flows from mRNA → Protein
- Occurs in ribosomes
- Involves tRNA, rRNA, amino acids, and enzymes
Site of Translation
| Organism | Site |
|---|---|
| Prokaryotes | Cytoplasm (ribosomes) |
| Eukaryotes | Cytoplasm (ribosomes) / Rough ER |
Components of Translation
| Component | Function |
|---|---|
| mRNA | Carries codons (genetic info) |
| tRNA | Brings specific amino acids to ribosome |
| rRNA | Structural & catalytic component of ribosome |
| Ribosome | Site of protein synthesis |
| Amino acids | Building blocks of proteins |
| Enzymes | Catalyze peptide bond formation |
Steps of Translation
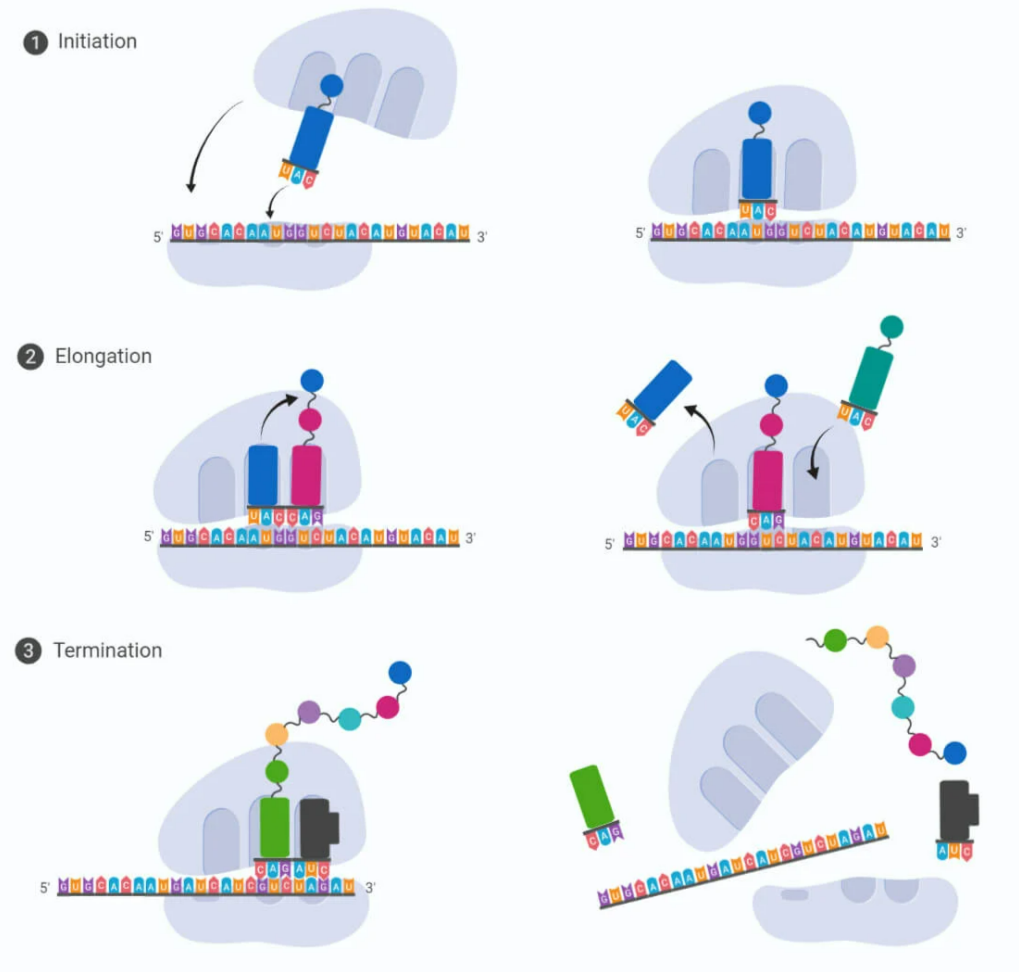
Step 1: Initiation
- Small ribosomal subunit binds to mRNA start codon (AUG)

- Initiator tRNA (Met) binds to AUG → initiation complex formed
- Large ribosomal subunit joins → functional ribosome
Step 2: Elongation
- tRNA brings amino acids to A site of ribosome
- Peptide bond forms between amino acids → polypeptide chain grows
- Ribosome moves along mRNA → next codon enters A site
- tRNA in P site releases amino acid after transfer
- Key Points:
- Peptide bonds catalyzed by peptidyl transferase (rRNA)
- Direction of translation: N-terminal → C-terminal
Step 3: Termination
- Ribosome reaches stop codon (UAA, UAG, UGA)
- Release factors bind → polypeptide chain released
- Ribosomal subunits dissociate
Important Features
| Feature | Description |
|---|---|
| Start codon | AUG → Methionine |
| Stop codon | UAA, UAG, UGA → no amino acid |
| Reading frame | Set by start codon, ensures correct codon grouping |
| Direction | mRNA read 5’→3’, protein synthesized N→C terminal |
Post-Translational Modifications
- Protein folding → functional 3D structure
- Glycosylation → sugar addition → glycoproteins
- Phosphorylation → activates/deactivates protein
- Cleavage → signal peptides removed for functional protein
🧾 Quick Recap
Transcription: DNA → RNA
Enzyme: RNA polymerase
Template strand: 3’→5’, RNA synthesized 5’→3’
RNA Types: mRNA, tRNA, rRNA
Eukaryotic modifications: 5’ cap, Poly-A tail, Splicing
Genetic Code: mRNA codons → amino acids
Triplet code: 1 codon = 1 amino acid
Start codon: AUG → Methionine
Stop codons: UAA, UAG, UGA
Degenerate: multiple codons for same amino acid
Translation: mRNA → Protein
Site: Ribosome (cytoplasm / RER)
Components: mRNA, tRNA, rRNA, Amino acids, Enzymes
Post-translational: Folding, glycosylation, phosphorylation, cleavage
Direction: mRNA 5’→3’, Protein N→C terminal
Lac Operon (Gene Expression and Regulation)
Introduction
Gene expression in prokaryotes is mainly controlled at the transcriptional level.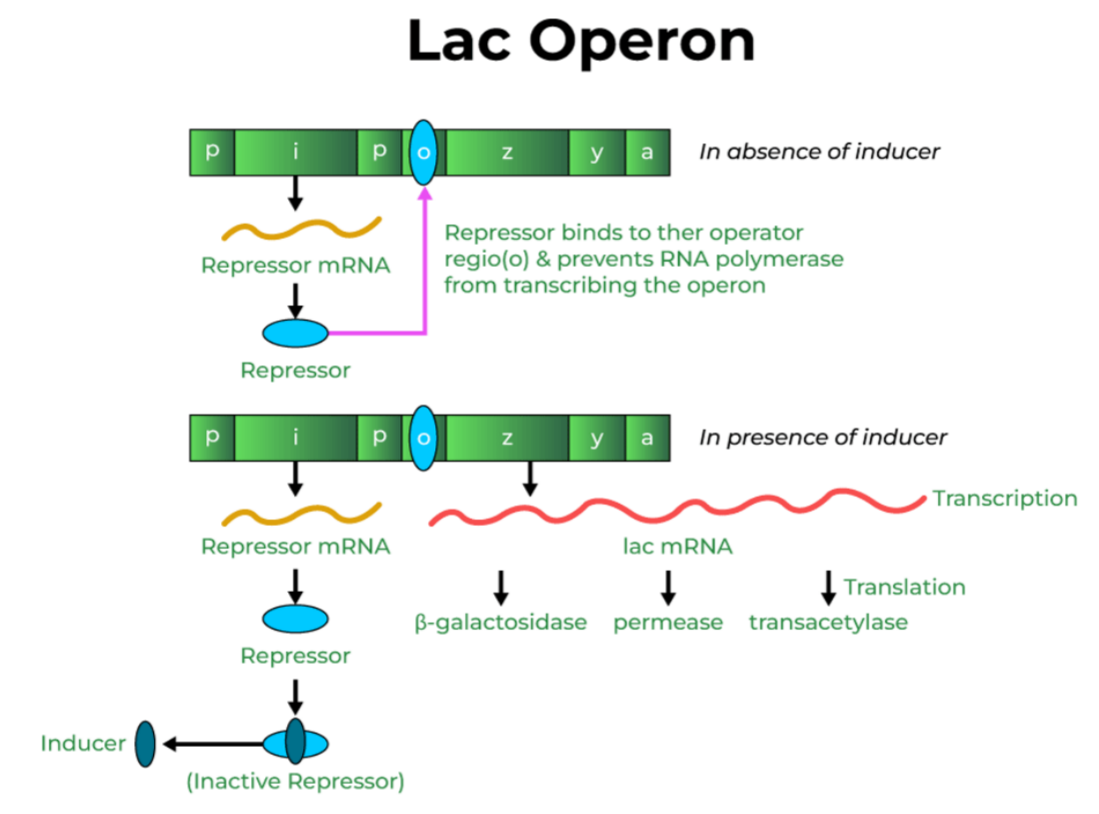
An operon is a group of genes regulated together under one promoter.
The Lac Operon in E. coli is the best-known model for gene regulation and is inducible.
What is Lac Operon?
Lac Operon is a cluster of genes in E. coli that gets switched ON only in the presence of lactose, allowing bacteria to break it down.
It is an example of an inducible operon because lactose acts as an inducer.
Purpose of Lac Operon
Helps bacteria use lactose as an energy source when glucose is absent.
Saves energy by not producing enzymes when lactose is absent.
🧩Components of Lac Operon
A. Structural Genes
| Gene | Function |
|---|---|
| lac Z | Makes beta-galactosidase (breaks lactose → glucose + galactose) |
| lac Y | Makes permease (helps lactose enter the cell) |
| lac A | Makes transacetylase (detoxification role) |
B. Regulatory Elements
| Element | Role |
|---|---|
| Promoter (P) | RNA polymerase binds here |
| Operator (O) | On-off switch controlled by repressor |
| Regulator gene (lacI) | Produces repressor protein |
🌿 How Lac Operon Works (ON-OFF Logic)
A. When lactose is absent → Operon OFF
- lacI produces active repressor
- Repressor binds to operator (O)
- RNA polymerase is blocked
- Structural genes not transcribed
No need to produce lactose-digesting enzymes if lactose is not there.
B. When lactose is present → Operon ON
- Lactose (actually allolactose) acts as inducer
- It binds to the repressor
- Repressor becomes inactive
- Operator is free
- RNA polymerase transcribes lacZ, lacY, lacA
Enzymes produced → lactose gets broken down
Lactose switches ON the system to digest itself.
🌿 Role of Glucose (Catabolite Repression)
Glucose is the preferred energy source.
When glucose is high:
- cAMP levels are low
- CAP–cAMP complex doesn’t form
- RNA polymerase binding becomes weak
- Lac operon is barely ON even if lactose is present
When glucose is low:
- cAMP level rises
- CAP–cAMP binds to promoter
- RNA polymerase binds strongly
- Operon strongly ON
Lactose ON
Glucose LOW
= Maximum expression
💡Summary Table
| Condition | Repressor | Lactose | Glucose | Operon Status |
|---|---|---|---|---|
| No lactose | Active | NO | Any | OFF |
| Lactose present | Inactive | YES | High | Weak ON |
| Lactose present | Inactive | YES | Low | Strong ON |
| Glucose present & no lactose | Active | NO | YES | OFF |
🌿 Important Points
- Lac operon is inducible (substrate induces gene expression)
- Lactose acts as natural inducer
- lacI is the regulator gene (makes repressor)
- Repressor binds operator
- CAP–cAMP enhances transcription when glucose is low
- Example of negative regulation (repressor blocks transcription)
🧾 Quick Recap
Lac operon = inducible operon in E. coli
lacZ, Y, A = enzymes for lactose breakdown
lacI makes repressor
Lactose binds repressor and removes it from operator
ON only when lactose present + glucose low
CAP–cAMP boosts RNA polymerase binding
Lac operon shows negative regulation and catabolite repression
Genome and Human Genome Project
🌿 1. Genome
1.1 Definition
Genome is the total genetic material of an organism, including all genes and non-coding sequences of DNA.
In simple words: the entire DNA content of a cell or organism.
Contains:
- Genes: coding regions → produce proteins
- Regulatory sequences and non-coding DNA → control gene expression and structural functions
1.2 Types of Genomes
| Type | Description | Examples |
|---|---|---|
| Nuclear Genome | DNA present in the nucleus | Human chromosomes (22 pairs autosomes + sex chromosomes) |
| Mitochondrial Genome | DNA in mitochondria | Human mitochondrial DNA (~16,569 bp) |
| Chloroplast Genome | DNA in chloroplasts (plants) | Arabidopsis, maize |
1.3 Human Genome
- Consists of ~3.2 billion base pairs
- Organized into 46 chromosomes (23 pairs)
- Contains ~20,000–25,000 protein-coding genes
- Majority (~98 percent) is non-coding DNA
Functions:
- Determines hereditary traits
- Provides blueprint for protein synthesis
- Non-coding DNA helps in regulation, chromosomal structure, and evolution
🧪 2. Human Genome Project (HGP)
2.1 Definition
The Human Genome Project was an international research effort to sequence the entire human DNA and map all human genes.
Goal: Understand the genetic makeup of humans.
Duration: 1990 to 2003
Led by: USA (NIH and DOE), with global collaboration
2.2 Objectives of HGP
- Sequence all 3.2 billion base pairs of human DNA
- Identify all human genes (~20,000 to 25,000)
- Store information in databases for research
- Improve tools for DNA sequencing and data analysis
- Address ethical, legal, and social issues (ELSI)
2.3 Achievements of HGP
| Achievement | Details |
|---|---|
| DNA Sequencing | ~3.2 billion base pairs sequenced |
| Gene Mapping | Identified ~20,500 protein-coding genes |
| Understanding Diseases | Mapped genetic basis of cancer, diabetes, cardiovascular disorders, etc. |
| Bioinformatics | Created databases and computational tools for DNA analysis |
| Personalized Medicine | Foundation for pharmacogenomics and targeted therapies |
| International Collaboration | Data shared globally with free access |
2.4 Applications of HGP
- Medical Genetics: Identifying disease-causing genes (example: BRCA1 and BRCA2)
- Pharmacogenomics: Tailoring medicines based on genetic makeup
- Forensics: DNA fingerprinting and paternity testing
- Evolutionary Biology: Comparing human genome with other species
- Gene Therapy: Correcting defective genes
2.5 Key Points
- HGP completed in 2003
- Only ~1 to 2 percent of genome codes for proteins
- Remaining ~98 percent is regulatory or non-coding
- Laid foundation for genomic medicine and personalized treatment
🧾 Quick Recap
Genome: Entire genetic material of an organism
Types: Nuclear, Mitochondrial, Chloroplast
Human Genome: 46 chromosomes, ~3.2 billion bp, ~20,000 to 25,000 genes
~98 percent is non-coding
Human Genome Project: 1990 to 2003
Goals: Sequence DNA and identify all genes
Achievements: Gene mapping, disease understanding, bioinformatics, personalized medicine
Applications: Genetics, gene therapy, forensics, evolution studies
DNA Fingerprinting and Protein Biosynthesis
🧬 DNA Fingerprinting
Definition
DNA fingerprinting (also called DNA profiling) is a technique used to identify individuals based on their unique DNA sequences.
Every person has a unique DNA pattern except identical twins.
It is widely used in forensics, paternity testing, and genetic research.
Principle
- DNA contains short tandem repeats (STRs) or variable number tandem repeats (VNTRs)
- These regions vary from person to person
- This variability becomes the basis for identification
Steps of DNA Fingerprinting
Sample Collection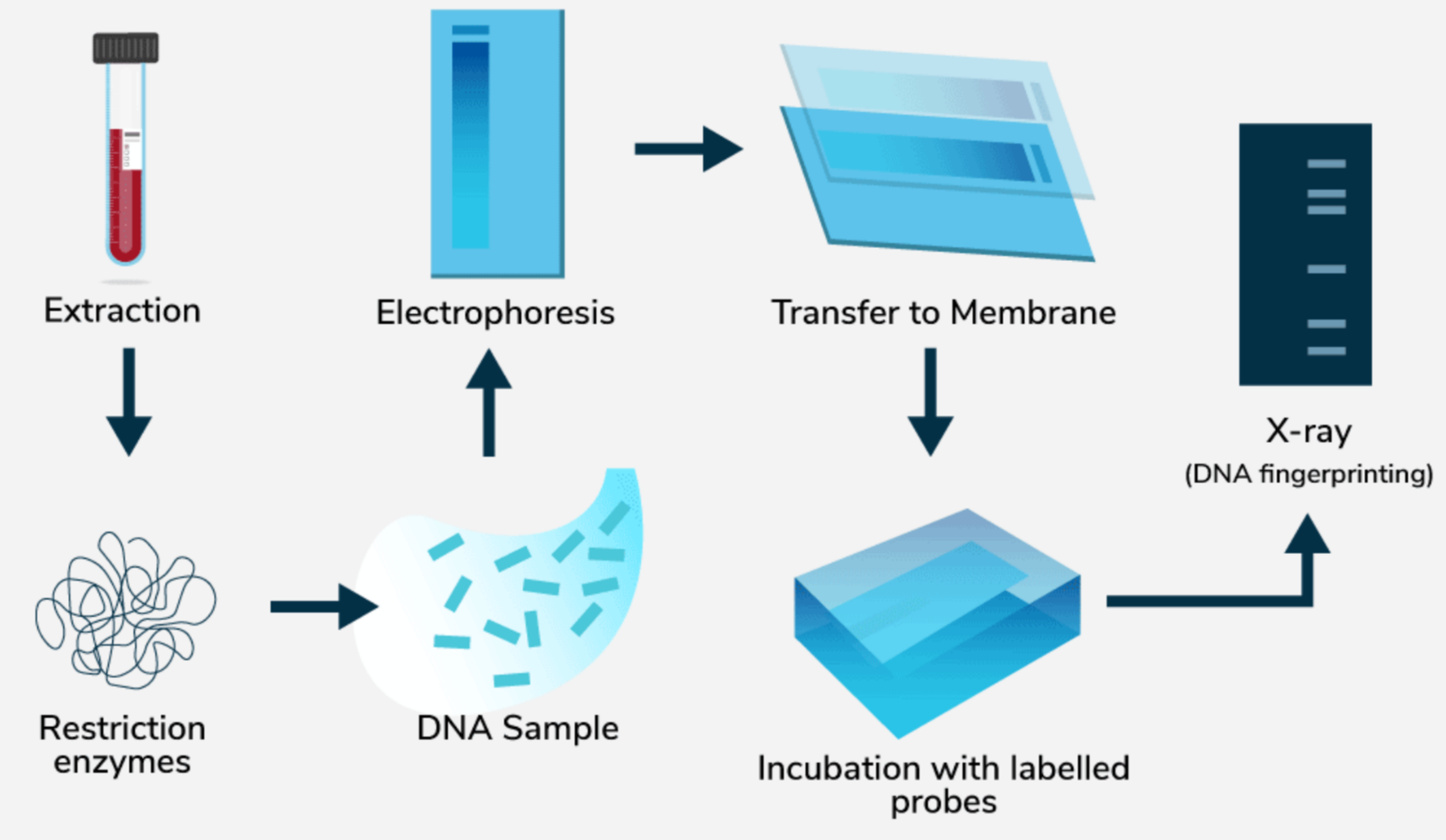
- Blood, hair roots, saliva, semen, skin cells
DNA Extraction
- DNA is purified from collected cells
DNA Cutting
- Restriction enzymes cut DNA at specific sequences
Amplification (PCR)
- PCR increases the quantity of selected DNA regions
Separation (Gel Electrophoresis)
- DNA fragments separated based on size and charge
Visualization and Comparison
- DNA band patterns are compared with references
- A unique pattern confirms identity
1.4 Applications
| Application | Example |
|---|---|
| Forensic Science | Crime scene investigation |
| Paternity Testing | Confirm biological relationships |
| Conservation Biology | Identify endangered species |
| Medical Genetics | Identify genetic disorders |
| Evolutionary Studies | Compare DNA across species |
1.5 Advantages
- Highly specific and reliable
- Effective even with small or degraded samples
- DNA pattern stays the same throughout life
🌿 Protein Biosynthesis
Definition
Protein biosynthesis is the process through which cells make proteins using the genetic information stored in DNA.
It includes two major steps: transcription and translation.
Steps of Protein Biosynthesis
A. Transcription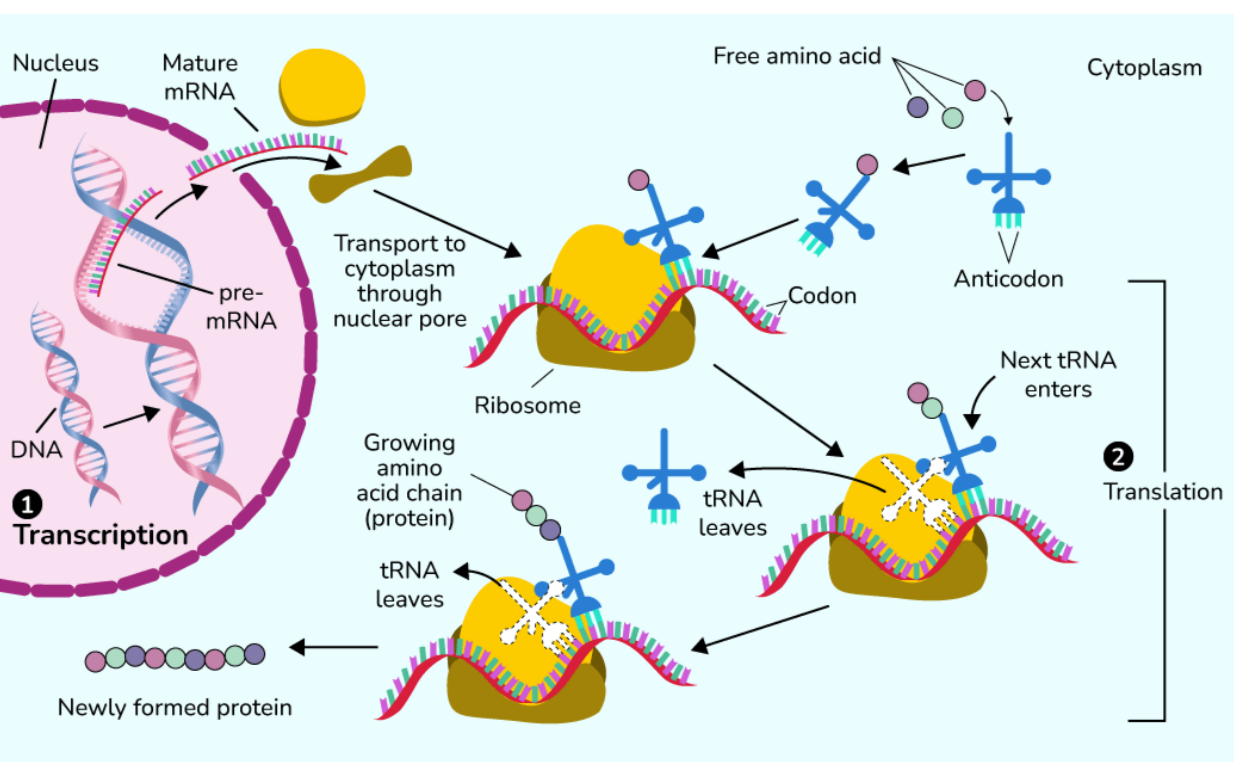
- Site: Nucleus in eukaryotes, cytoplasm in prokaryotes
- Enzyme: RNA polymerase
Process:
- Initiation: RNA polymerase binds to promoter
- Elongation: RNA strand grows in 5’ to 3’ direction
- Termination: RNA transcript is released
RNA Processing (Eukaryotes)
- 5’ capping
- 3’ poly A tail addition
- Splicing to remove introns
B. Translation
- Site: Ribosomes (cytoplasm or rough ER)
- Components: mRNA, tRNA, rRNA, ribosome
Process:
- Initiation: Ribosome recognizes start codon (AUG)
- Elongation: tRNA brings amino acids and peptide bonds form
- Termination: Stop codon reached, polypeptide released
Genetic Code
- Triplet code: 3 bases make 1 codon
- Start codon: AUG (methionine)
- Stop codons: UAA, UAG, UGA
- Degenerate: multiple codons can code for same amino acid
Post Translational Modifications
- Protein folding for functional 3D shape
- Glycosylation: sugar addition
- Phosphorylation: activity regulation
- Cleavage: removal of signal peptides
🧾 Quick Recap
DNA Fingerprinting: Unique DNA pattern for each individual
Steps: Sample → Extraction → Enzymes → PCR → Gel → Compare
Applications: Forensics, paternity testing, conservation, medical genetics
Protein Biosynthesis: DNA to mRNA to protein
Transcription: Nucleus, RNA polymerase, capping, tailing, splicing
Translation: Ribosomes, tRNA, rRNA, peptide bond formation
Genetic Code: Triplet code, AUG start, stop codons, degeneracy
Modifications: Folding, glycosylation, phosphorylation